2.7.1: Control of Metabolism Through Enzyme Regulation
- Page ID
- 8810
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Explain the effect of an enzyme on chemical equilibrium
Control of Metabolism Through Enzyme Regulation
Cellular needs and conditions vary from cell to cell and change within individual cells over time. For example, a stomach cell requires a different amount of energy than a skin cell, fat storage cell, blood cell, or nerve cell. The same stomach cell may also need more energy immediately after a meal and less energy between meals.

A cell’s function is encapsulated by the chemical reactions it can carry out. Enzymes lower the activation energies of chemical reactions; in cells, they promote those reactions that are specific to the cell’s function. Because enzymes ultimately determine which chemical reactions a cell can carry out and the rate at which they can proceed, they are key to cell functionality.
Competitive and Noncompetitive Inhibition
The cell uses specific molecules to regulate enzymes in order to promote or inhibit certain chemical reactions. Sometimes it is necessary to inhibit an enzyme to reduce a reaction rate, and there is more than one way for this inhibition to occur. In competitive inhibition, an inhibitor molecule is similar enough to a substrate that it can bind to the enzyme’s active site to stop it from binding to the substrate. It “competes” with the substrate to bind to the enzyme.
In noncompetitive inhibition, an inhibitor molecule binds to the enzyme at a location other than the active site (an allosteric site). The substrate can still bind to the enzyme, but the inhibitor changes the shape of the enzyme so it is no longer in optimal position to catalyze the reaction.

Allosteric Inhibition and Activation
In noncompetitive allosteric inhibition, inhibitor molecules bind to an enzyme at the allosteric site. Their binding induces a conformational change that reduces the affinity of the enzyme’s active site for its substrate. The binding of this allosteric inhibitor changes the conformation of the enzyme and its active site, so the substrate is not able to bind. This prevents the enzyme from lowering the activation energy of the reaction, and the reaction rate is reduced.
However, allosteric inhibitors are not the only molecules that bind to allosteric sites. Allosteric activators can increase reaction rates. They bind to an allosteric site which induces a conformational change that increases the affinity of the enzyme’s active site for its substrate. This increases the reaction rate.
Cofactors and Coenzymes
Many enzymes only work if bound to non-protein helper molecules called cofactors and coenzymes. Binding to these molecules promotes optimal conformation and function for their respective enzymes. These molecules bind temporarily through ionic or hydrogen bonds or permanently through stronger covalent bonds.
Cofactors are inorganic ions such as iron (Fe2+) and magnesium (Mg2+). For example, DNA polymerase requires a zinc ion (Zn2+) to build DNA molecules. Coenzymes are organic helper molecules with a basic atomic structure made up of carbon and hydrogen. The most common coenzymes are dietary vitamins. Vitamin C is a coenzyme for multiple enzymes that take part in building collagen, an important component of connective tissue. Pyruvate dehydrogenase is a complex of several enzymes that requires one cofactor and five different organic coenzymes to catalyze its chemical reaction. The availability of various cofactors and coenzymes regulates enzyme function.

Enzyme Compartmentalization
In eukaryotic cells, molecules such as enzymes are usually compartmentalized into different organelles. This organization contributes to enzyme regulation because certain cellular processes are contained in separate organelles. For example, the enzymes involved in the later stages of cellular respiration carry out reactions exclusively in the mitochondria. The enzymes involved in the digestion of cellular debris and foreign materials are located within lysosomes.
Feedback Inhibition in Metabolic Pathways
Feedback inhibition is when a reaction product is used to regulate its own further production. Cells have evolved to use feedback inhibition to regulate enzyme activity in metabolism, by using the products of the enzymatic reactions to inhibit further enzyme activity. Metabolic reactions, such as anabolic and catabolic processes, must proceed according to the demands of the cell. In order to maintain chemical equilibrium and meet the needs of the cell, some metabolic products inhibit the enzymes in the chemical pathway while some reactants activate them.
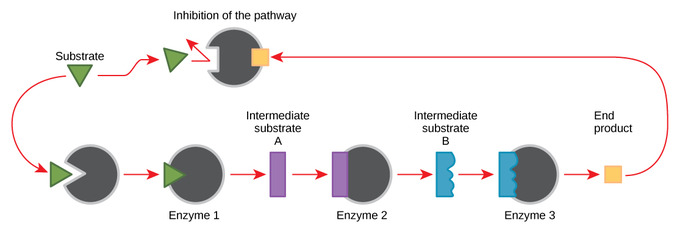
The production of both amino acids and nucleotides is controlled through feedback inhibition. For an example of feedback inhibition, consider ATP. It is the product of the catabolic metabolism of sugar (cellular respiration), but it also acts as an allosteric regulator for the same enzymes that produced it. ATP is an unstable molecule that can spontaneously dissociate into ADP; if too much ATP were present, most of it would go to waste. This feedback inhibition prevents the production of additional ATP if it is already abundant. However, while ATP is an inhibitor, ADP is an allosteric activator. When levels of ADP are high compared to ATP levels, ADP triggers the catabolism of sugar to produce more ATP.
Key Points
- In competitive inhibition, an inhibitor molecule competes with a substrate by binding to the enzyme ‘s active site so the substrate is blocked.
- In noncompetitive inhibition (also known as allosteric inhibition), an inhibitor binds to an allosteric site; the substrate can still bind to the enzyme, but the enzyme is no longer in optimal position to catalyze the reaction.
- Allosteric inhibitors induce a conformational change that changes the shape of the active site and reduces the affinity of the enzyme’s active site for its substrate.
- Allosteric activators induce a conformational change that changes the shape of the active site and increases the affinity of the enzyme’s active site for its substrate.
- Feedback inhibition involves the use of a reaction product to regulate its own further production.
- Inorganic cofactors and organic coenzymes promote optimal enzyme orientation and function.
- Vitamins act as coenzymes (or precursors to coenzymes) and are necessary for enzymes to function.
Key Terms
- coenzyme: An organic molecule that is necessary for an enzyme to function.
- allosteric site: A site other than the active site on an enzyme.
- cofactor: An inorganic molecule that is necessary for an enzyme to function.


