18.3: Phenotypes and Genotypes
- Page ID
- 70541
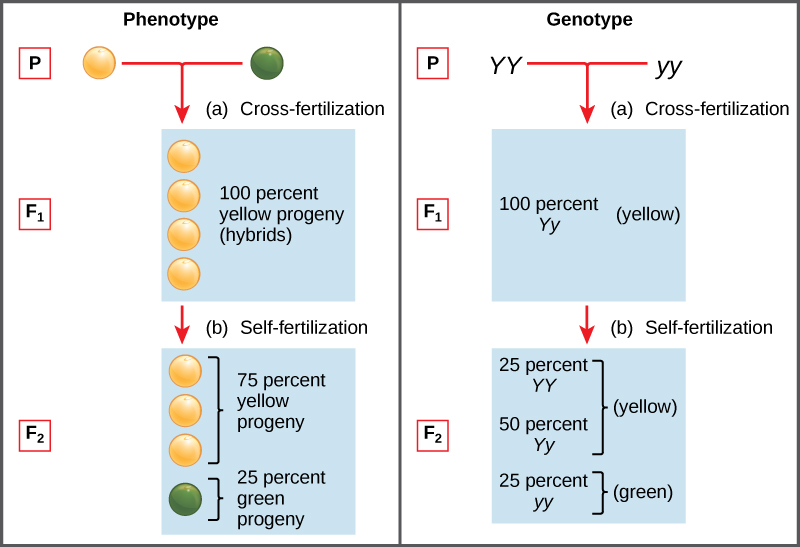
Two alleles for a given gene in a diploid organism are expressed and interact to produce physical characteristics. The observable traits expressed by an organism are referred to as its phenotype. An organism’s underlying genetic makeup, consisting of both the physically visible and the non-expressed alleles, is called its genotype. Mendel’s hybridization experiments demonstrate the difference between phenotype and genotype. For example, the phenotypes that Mendel observed in his crosses between pea plants with differing traits are connected to the diploid genotypes of the plants in the P, F1, and F2 generations. We will use a second trait that Mendel investigated, seed color, as an example. Seed color is governed by a single gene with two alleles. The yellow-seed allele is dominant and the green-seed allele is recessive. When true-breeding plants were cross-fertilized, in which one parent had yellow seeds and one had green seeds, all of the F1 hybrid offspring had yellow seeds. That is, the hybrid offspring were phenotypically identical to the true-breeding parent with yellow seeds. However, we know that the allele donated by the parent with green seeds was not simply lost because it reappeared in some of the F2 offspring (Figure \(\PageIndex{1}\)). Therefore, the F1 plants must have been genotypically different from the parent with yellow seeds.
The P plants that Mendel used in his experiments were each homozygous for the trait he was studying. Diploid organisms that are homozygous for a gene have two identical alleles, one on each of their homologous chromosomes. The genotype is often written as YY or yy, for which each letter represents one of the two alleles in the genotype. The dominant allele is capitalized and the recessive allele is lower case. The letter used for the gene (seed color in this case) is usually related to the dominant trait (yellow allele, in this case, or “Y”). Mendel’s parental pea plants always bred true because both produced gametes carried the same allele. When P plants with contrasting traits were cross-fertilized, all of the offspring were heterozygous for the contrasting trait, meaning their genotype had different alleles for the gene being examined. For example, the F1 yellow plants that received a Y allele from their yellow parent and a y allele from their green parent had the genotype Yy.
Law of Dominance
Our discussion of homozygous and heterozygous organisms brings us to why the F1 heterozygous offspring were identical to one of the parents, rather than expressing both alleles. In all seven peaplant characteristics, one of the two contrasting alleles was dominant, and the other was recessive. Mendel called the dominant allele the expressed unit factor; the recessive allele was referred to as the latent unit factor. We now know that these so-called unit factors are actually genes on homologous chromosomes. For a gene that is expressed in a dominant and recessive pattern, homozygous dominant and heterozygous organisms will look identical (that is, they will have different genotypes but the same phenotype), and the recessive allele will only be observed in homozygous recessive individuals (Table \(\PageIndex{1}\)).
Table \(\PageIndex{1}\): Correspondence between Genotype and Phenotype for a Dominant Recessive Characteristic.
| Homozygous | Heterozygous | Homozygous | |
|---|---|---|---|
| Genotype | YY | Yy | yy |
| Phenotype | yellow | yellow | green |
Mendel’s law of dominance states that in a heterozygote, one trait will conceal the presence of another trait for the same characteristic. For example, when crossing true-breeding violet-flowered plants with true-breeding white-flowered plants, all of the offspring were violet-flowered, even though they all had one allele for violet and one allele for white. Rather than both alleles contributing to a phenotype, the dominant allele will be expressed exclusively. The recessive allele will remain latent, but will be transmitted to offspring in the same manner as that by which the dominant allele is transmitted. The recessive trait will only be expressed by offspring that have two copies of this allele (Figure \(\PageIndex{2}\)), and these offspring will breed true when self-crossed.

References
Unless otherwise noted, images on this page are licensed under CC-BY 4.0 by OpenStax.
OpenStax, Biology. OpenStax CNX. May 27, 2016 http://cnx.org/contents/GFy_h8cu@10....ics-and-Traits

