12.1: The lac Operon
- Page ID
- 4150
Early insights into mechanisms of transcriptional regulation came from studies of E. coli by researchers Francois Jacob & Jacques Monod. In E. coli, and many other bacteria, genes encoding several different proteins may be located on a single transcription unit called an operon. The genes in an operon share the same transcriptional regulation, but are translated individually. Eukaryotes generally do not group genes together as operons (exception is C. elegans and a few other species).
Basic lac Operon structure
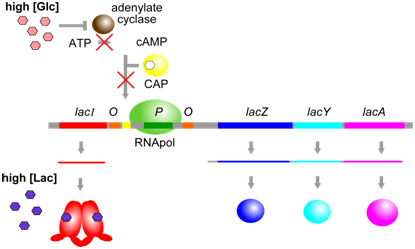
E. coli encounters many different sugars in its environment. These sugars, such as lactose and glucose, require different enzymes for their metabolism. Three of the enzymes for lactose metabolism are grouped in the lac operon: lacZ, lacY, and lacA (Figure \(\PageIndex{1}\)). LacZ encodes an enzyme called β-galactosidase, which digests lactose into its two constituent sugars: glucose and galactose. lacY is a permease that helps to transfer lactose into the cell. Finally, lacA is a trans-acetylase; the relevance of which in lactose metabolism is not entirely clear. Transcription of the lac operon normally occurs only when lactose is available for it to digest. Presumably, this avoids wasting energy in the synthesis of enzymes for which no substrate is present. A single mRNA transcript includes all three enzyme-coding sequences and is called polycistronic. A cistron is equivalent to a gene.

cis- and transRegulators
In addition to the three protein-coding genes, the lac operon contains short DNA sequences that do not encode proteins, but are instead binding sites for proteins involved in transcriptional regulation of the operon. In the lac operon, these sequences are called P (promoter), O (operator), and CBS (CAP-binding site). Collectively, sequence elements such as these are called cis-elements because they must be located on the same piece of DNA as the genes they regulate. On the other hand, the proteins that bind to these cis-elements are called trans-regulators because (as diffusible molecules) they do not necessarily need to be encoded on the same piece of DNA as the genes they regulate.
lacI is an allosterically regulated repressor
One of the major trans-regulators of the lac operon is encoded by lacI. Four identical molecules of lacI proteins assemble together to form a homotetramer called a repressor (Figure \(\PageIndex{2}\)). This repressor binds to two operator sequences adjacent to the promoter of the lac operon. Binding of the repressor prevents RNA polymerase from binding to the promoter (Figure \(\PageIndex{3}\)). Therefore, the operon will not be transcribed when the operator is occupied by a repressor.

Besides its ability to bind to specific DNA sequences at the operator, another important property of the lacI protein is its ability to bind to lactose. When lactose is bound to lacI, the shape of the protein changes in a way that prevents it from binding to the operator. Therefore, in the presence of lactose, RNA polymerase is able to bind to the promoter and transcribe the lac operon, leading to a moderate level of expression of the lacZ, lacY, and lacA genes. Proteins such as lacI that change their shape and functional properties after binding to a ligand are said to be regulated through an allosteric mechanism. The role of lacI in regulating the lac operon is summarized in Figure \(\PageIndex{4}\).

CAP is an allosteric activator of the lac operon
A second aspect of lac operon regulation is conferred by a trans-factor called cAMP binding protein (CAP, Figure \(\PageIndex{4}\)). CAP is another example of an allosterically regulated trans-factor. Only when the CAP protein is bound to cAMP can another part of the protein bind to a specific cis-element within the lac promoter called the CAP binding sequence (CBS). CBS is located very close to the promoter (P). When CAP is bound to at CBS, RNA polymerase is better able to bind to the promoter and initiate transcription. Thus, the presence of cAMP ultimately leads to a further increase in lac operon transcription.

The physiological significance of regulation by cAMP becomes more obvious in the context of the following information. The concentration of cAMP is inversely proportional to the abundance of glucose: when glucose concentrations are low, an enzyme called adenylate cyclase is able to produce cAMP from ATP. Evidently, E. coli prefers glucose over lactose, and so expresses the lac operon at high levels only when glucose is absent and lactose is present. This provides another layer of logical control of lac operon expression: only in the presence of lactose, and in the absence of glucose is the operon expressed at its highest levels.