28.13: Regulation of the Cell Cycle by Protein Kinases
- Page ID
- 15122
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Below is a set of learning goals designed to guide junior and senior biochemistry majors through the key concepts and mechanisms involved in cell cycle regulation by cyclin-dependent kinases (CDKs), cyclins, and their associated regulatory systems:
-
Outline the Mammalian Cell Cycle:
- Describe the four major phases (G1, S, G2, and M) and the decision points (G0, differentiation, re-entry into the cycle) that regulate cell division.
- Explain how cells prepare for DNA replication and mitosis through the synthesis of macromolecules and cell growth.
-
Identify Core Regulatory Players:
- Define the roles of cyclin-dependent kinases (CDKs), cyclins, and CDK inhibitors (CKIs) in controlling cell cycle progression.
- Explain the specific functions of key CDK-cyclin complexes (e.g., CDK3/cyclin C, CDK4/6/cyclin D, CDK2/cyclin E, CDK2/cyclin A, CDK1/cyclin B) in transitioning between cell cycle phases.
-
Understand pRb Phosphorylation and E2F Release:
- Describe how phosphorylation of the retinoblastoma protein (pRb) by CDK-cyclin complexes leads to the release of E2F transcription factors, driving the G1/S transition.
-
Interpret Cyclin Expression and CDK Oscillations:
- Summarize the coordinated expression of cyclins throughout the cell cycle and how their oscillatory levels regulate CDK activity.
- Analyze graphical representations of CDK-cyclin oscillations and discuss how these oscillations correlate with cell cycle events.
-
Differentiate CDKs with Additional Functions:
- Compare classical CDKs involved directly in cell cycle progression with those that have roles in transcription, DNA repair, and other cellular processes.
- Describe how transcription-related CDKs (e.g., CDK7, CDK8, CDK9, CDK12/13) regulate RNA polymerase II and influence gene expression.
-
Examine Structural Features of CDKs and Cyclins:
- Identify key structural motifs in CDKs (e.g., the N-terminal lobe, C-terminal lobe, C-helix, hinge region, activation loop, and glycine-rich loop) and discuss their roles in catalytic activity.
- Explain how cyclin binding induces conformational changes in CDKs that are critical for their activation.
-
Understand Substrate Recognition Mechanisms:
- Describe how specific substrate motifs (e.g., the S/T-PX(K/R) and RXL motifs) in target proteins enable selective phosphorylation by CDK-cyclin complexes.
- Explain the role of distal docking sites (e.g., the MRAIL motif in cyclins) in enhancing substrate specificity.
-
Discuss the Role of CDK-Activating Kinases (CAK):
- Explain how CAK (a complex of CDK7, cyclin H, and MAT1) phosphorylates the activation loop of CDKs, thereby enhancing their kinase activity.
- Describe the dual role of CAK in both cell cycle regulation and transcription initiation.
-
Apply Mathematical Modeling to Cell Cycle Oscillations:
- Summarize the concept of using mass action kinetics and Hill equations to model feedback loops in the CDK network.
- Discuss how negative feedback (via the APC complex) and multi-species models (incorporating CDK1, APC, and Plk1) contribute to the oscillatory behavior observed in cell cycle regulation.
-
Integrate Experimental and Computational Approaches:
- Understand how structural models (e.g., iCn3D models of CDK2, cyclin A, and CAK) support our knowledge of CDK-cyclin interactions.
- Evaluate the role of computational simulations (such as those performed in VCell) in testing kinetic models and predicting oscillatory dynamics in cell cycle regulators.
By achieving these learning goals, students will gain a comprehensive understanding of how CDK/cyclin complexes regulate the cell cycle, the structural and kinetic basis for their activity, and the integration of experimental and computational methods in studying cell cycle control.
This section is an integration of materials as referenced, with significant modifications and additions.
Aleem and Arceci. Targeting cell cycle regulators in hematologic malignancies. Article in Frontiers in Cell and Developmental Biology 2015. DOI: 10.3389/fcell.2015.00016. Creative Commons Attribution 4.0 International
Introduction
For a cell to undergo successful division, it has to perform four key tasks in a highly ordered fashion. First, a preparatory synthetic phase (G1) results in increased cell size in anticipation of DNA replication (S phase). Cells then proceed through the G2-phase to prepare to equally segregate duplicated DNA (M phase) and finally divide into two equal daughter cells. From G1, a cell can exit the cell cycle and enter a state of quiescence (G0), undergo differentiation, or re-enter the cell cycle to proliferate in response to mitogenic signals.
The core molecular machinery controlling the mammalian cell cycle consists of a family of serine/threonine protein kinases called cyclin-dependent kinases (CDKs). These catalytic subunits are activated in most cases by association with cyclin regulatory subunits. The activity of CDK/cyclin complexes is further regulated by CDK inhibitors (CKIs), phosphorylation and dephosphorylation, ubiquitin-mediated degradation, transcriptional regulation, substrate recognition, and subcellular localization. The family of CDKs/cyclins/CKIs contains more than 30 members. They are implicated in essential cellular functions such as transcription, DNA damage repair, epigenetic regulation, metabolism, proteolytic degradation, stem cell self-renewal, neuronal functions, and spermatogenesis. Figure \(\PageIndex{1}\) shows the cell cycle and the involvement of CDKs/cyclins at key points.
CDK3/cyclin C drives cell cycle entry from G0. CDK4/6/cyclin D complexes initiate phosphorylation of the retinoblastoma protein (pRb), and they sequester p21Cip1 and p27kip1 (not shown), which are both inhibitors of CDK2, thus promoting the activation of CDK2/cyclin E complex. In late G1, the CDK2/cyclin E complex completes phosphorylation and inactivation of pRb, which releases the E2F transcription factors, and the G1/S transition takes place. DNA replication takes place in the S phase. CDK2/cyclin A complex regulates progression through the S phase, and CDK1/cyclin A complex through the G2 phase in preparation for mitosis (M). Mitosis is initiated by CDK1/cyclin B complex (which will be modeled at this section's end). The activity of CDK1/cyclin B is tightly regulated by activating phosphorylation by the CDK-activating kinase CAK (a heterodimer of cyclin H-CDK7-MAT1) and inhibitory phosphorylations by Wee1 and Myt1 on Tyr15 and Thr14 (not shown). Some specific CDK4/CDK6 pharmacological inhibitors are also shown
CDKs with Direct Functions in Cell Cycle Regulation
The classical CDKs that directly regulate the mammalian cell cycle in complexes with cyclin subunits include CDK3, CDK4, CDK6, CDK2, and CDK1. CDK3 promotes cell cycle entry from quiescence in association with cyclin C. CDK8 has also been suggested to play a role in cell cycle entry from G0 and in the G1/S transition. In its simplest model, the mammalian cell cycle proceeds as follows:
- In early G1, CDK4/CDK6 in complex with cyclin D receive mitogenic signals that activate cell cycle entry, as shown in Figure \(\PageIndex{1}\). Key signaling events include the initiation of retinoblastoma protein (pRb) phosphorylation and the sequestration of p21Cip1 and p27kip1, which are both inhibitors of CDK2, thus promoting the activation of CDK2/ cyclin E complex. In late G1, CDK2 in complex with cyclin E completes the phosphorylation and inactivation of pRb, releasing the E2F transcription factors. E2F promotes transcription of cyclin E, which is necessary for the G1/S transition.
- Progression through the S phase is mediated by CDK2/cyclin A complex.
- CDK1/cyclin B complexes then initiate mitosis. We will model the regulation of CDK1 later in this section. CDK1/cyclin A complexes contribute to the preparation for mitosis in the G2 phase. The activity of CDK1/cyclin B is tightly regulated by activating phosphorylation by the CDK-activating kinase (CAK) (a heterodimer of cyclin H and CDK7) and inhibitory phosphorylations by WEE-1 and Myt1 on Tyr15 and Thr14. Mitosis starts after WEE-1 is degraded and CDC25C phosphatase releases the inhibitory phosphorylation on CDK1/cyclin B.
The cyclins are also expressed in a coordinated fashion throughout the cell cycle. Figure \(\PageIndex{2}\) shows the cyclin expression cycle. The timing of expression is consistent with the explanations above.
Figure \(\PageIndex{3}\) shows a graph showing multiple progressions through the cell cycle.
Figure \(\PageIndex{3}\): Sustained oscillations of the CDK network in mammalian cells. Gérard and Goldbeter, Front. Physiol., 02 November 2012
Sec. Systems Biology Archive. https://doi.org/10.3389/fphys.2012.00413. Creative Commons Attribution License
The time evolution of cyclin D/Cdk4–6 (in black), cyclin E/Cdk2 (in blue), cyclin A/Cdk2 (in green), and cyclin B/Cdk1 (in red) is shown in the presence of a suprathreshold level of growth factor. Cyclin D/Cdk4–6 is the total active form of the kinases, which is composed of cyclin D/Cdk4–6 and also the complex formed by cyclin D/Cdk4–6 and p21/p27.
Note again the oscillatory nature of the cyclin B/Cdk1 complex, which will be explored at the end of this section.
CDKs with Transcriptional and Other Functions
In addition to their direct role in the mitotic cell cycle regulation, some classical CDK/cyclin complexes have essential functions in meiosis, such as CDK2, in transcription, and/or DNA repair. Other CDKs act by activating the classical CDKs, such as CDK7/cyclin H (CAK) and the related CDK20, also known as cell cycle-related kinase (CCRK). Some CDKs function mainly in influencing transcription by phosphorylating the carboxy-terminal domain (CTD) of ribonucleic acid (RNA) polymerase II (RNA pol II). This phosphorylation also serves as a platform for RNA processing and chromatin regulation.
CDKs with important transcriptional roles include the CDK7/cyclin H/MAT1 complex, a component of the basal transcription factor TFIIH, which facilitates transcriptional initiation. CDK8/cyclin C, in addition to its role in transcription, is also involved in the Wnt/β-catenin pathway and inhibition of lipogenesis. Cyclin C can recruit CDK8 or CDK19 to the CDK8 module of the Mediator complex, which can function as a positive or negative transcription regulator by RNA pol II. CDK3/cyclin C also plays a role in NHEJ-mediated DNA damage repair. While CDK9 in complex with cyclin T forms the phospho-transcription elongation factor b (p-TEFb) and promotes transcriptional elongation, CDK9 also functions in the DNA damage response when complexed with cyclin K. CDK10/cyclin M phosphorylates the Ets2 transcription factor and positively controls its degradation by the proteasome. Ets2 plays a key role in cancer and development. CDK11/cyclin L controls the RNA pol II mediator complex assembly. CDK12 and CDK13 in complex with cyclin K control RNA pol II transcription, and CDK12/cyclin K controls DNA damage response.
Structure of CDKs–cyclins
Open Biology. Wood and Jane A. Endicott (2018) https://doi.org/10.1098/rsob.180112. Creative Commons Attribution License http://creativecommons.org/licenses/by/4.0/,
The structures of inactive cyclin-free kinases are very similar but vary at the N-terminal and C-terminal ends. Figure \(\PageIndex{4}\) shows an interactive iCn3D model of the prototypical active human cyclin-dependent kinase 2 with a bound ATP (1HCK).
.png?revision=1&size=bestfit&width=272&height=299)
The model shows that CDK2 has structural features shown in all the kinases we have studied previously:
- a smaller N-terminal lobe (light cyan) and larger C-terminal lobe (light magenta) in between which ATP binds (along with Mg2+).
- the C-helix (residues 45 – 55, purple) contains a conserved Glu. It forms an interaction with and helps position a key lysine in the active site, which facilitates ATP binding and transition state stabilization;
- hinge (residues 80 – 84, yellow),
- activation loop (residues 145– 172, red), which contains T160 (sticks, CPK colors, labeled) that becomes phosphorylated on activation by yet another kinase called CDK-activation kinase (CAK). When T160 is phosphorylated, the kinase binds to cyclin A. The loop starts and ends with the conserved residues DFG and APE, respectively.
- Not highlighted in the model is a conserved conformationally flexible glycine-rich region (residue 12-16) with the motif GXGXXG
In the inactive form, the N-terminal end of the activation loop has a short alpha helix that prevents the C-helix from adopting the correct position for catalysis. Activation requires movement of the C-helix, allowing the Glu in the C-helix to position the active site Lys.
CDK2–cyclin A activation
Cyclin A's binding to CDK2 activates it through the repositioning of the C-helix and the activation loop. When CDK2 is phosphorylated and bound to cyclin A, there is a large shift in the C-helix, allowing the interaction of the C-helix Glu with the active site Lys.
First, let's look at the structure of cyclins. Each cyclin has a unique sequence and structural features that allow it to interact with specific CDKs and associated proteins. However, they all have a conserved "cyclin box" structure containing about 100 amino acids.
Figure \(\PageIndex{5}\) shows an interactive iCn3D model of bovine cyclin A (1VIN).
%25C2%25A0.png?revision=1&size=bestfit&width=336&height=249)
Cyclin A has two linked cyclin box folds, each containing around 100 amino acids and five helices. They interact with the more disordered parts of unphosphorylated CDK2, which results in some low activity levels. Cyclin binding causes a large movement of the C-helix, enabling the Glu -- Lys interaction. Phosphorylation of T160 leads to the repositioning of the activation loop.
Figure \(\PageIndex{6}\) shows an interactive iCn3D model of Phosphorylated cyclin-dependent kinase 2 bound to cyclin A (1JST)
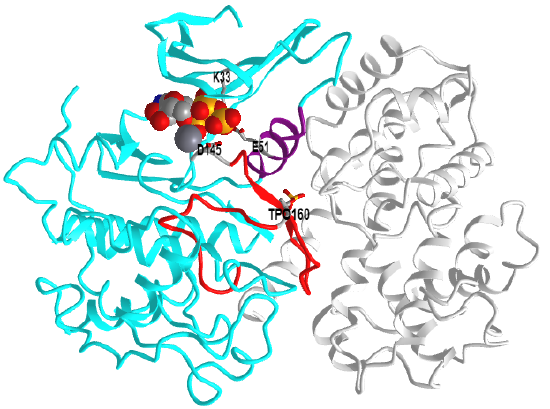
CDK2 is shown in cyan and cyclin A in gray. Here are some structural features represented in the model.
- the C-helix (residues 45 – 55, purple),
- activation loop (residues 145– 172, red), which contains pT160 (sticks, CPK colors, labeled)
- the catalytic "triad" Lys33, Glu51, and Asp145
Figure \(\PageIndex{7}\) shows an animation of structural changes in just CDK2 when "apo"-CDK2 (without bound cyclin A, pdbID 1HCK) binds cyclin A (1JST).
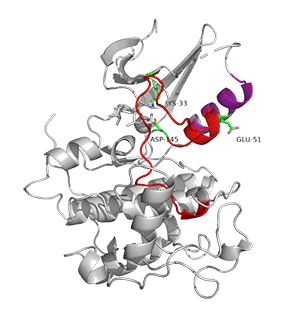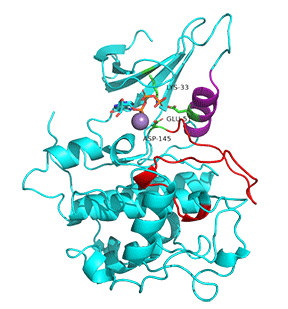
Gray represents the structure of CDK2 in the absence of cyclin A. The structure of just CDK2 in the presence of cyclin A is shown in cyan. Large shifts in the C-helix (purple) and activation loop (red) occur on binding cyclin A.
Cyclin partners of CDK1 and CDK2
CDK1 is the closest member of the CDK family to CDK2, for which structures of the cyclin-free and authentic cyclin-bound forms can also be compared.
Depending on cyclin availability and concentration, CDK2 can bind cyclin A, B (if CDK1 expression is knocked down), and E (see Figure \(\PageIndex{1}\). The binding interface between CDK2 and the cyclins is quite large compared to that between CDK1 and the cyclins. Three large aromatic side chains (Y170, Y177, and Y258) are conserved in the binding interface. The corresponding amino acids in cyclin E are smaller (N112, I119, and L202).
The binding interface between CDK1 and the cyclins is smaller so it appears that it might preferentially interact with cyclins A and B to gain binding affinity through the more robust interactions with the aromatic groups in the interface in the CDK2: Cyclin A and CDK2:cyclin B complexes.
A comparison of the CDK1–cyclin B and CDK2–cyclin A/B/E structures also highlights the potential for these closely related CDKs to be differentially regulated by reversible phosphorylation. The antagonistic activities of Wee1/Myt1 kinases and Cdc25 phosphatases regulate the phosphorylation status of the CDK glycine-rich loop (defined by the GXGXXG motif, residues 11–16 in CDK2). The structure of CDK2–cyclin A phosphorylated on Y15 illustrates how phosphorylation promotes a glycine loop structure that antagonizes both peptide substrate binding and the ATP conformation required for catalysis. The flexibility of the glycine-rich loop is compatible with a model in which the phosphorylated Y15 side chain is solvent-exposed and accessible to both kinases and phosphatases. CDK1 is also regulated by active-site phosphorylation, and the conserved nature of the structure in this region suggests that the mechanism of inhibition is also conserved.
CDK substrate recognition
Local and distal sequence motifs must be used to confer specificity to the binding of specific cyclins and other proteins to specific CDKs. One interesting example is examining the structure of a phospho-CDK2 Cyclin A in complex with a peptide substrate derived from the protein CDC6. Figure \(\PageIndex{8}\) shows an interactive iCn3D model of Phospho-CDK2:Cyclin A complex with a peptide containing both the substrate and recruitment sites of CDC6 (2CCI)
.png?revision=1&size=bestfit&width=351&height=307)
The color coding is the same as the models above:
- CDK2 is shown in cyan and cyclin A in gray.
- the C-helix (residues 45 – 55, purple),
- activation loop (residues 145– 172, red), which contains pT160 (sticks, CPK colors, labeled)
- the catalytic "triad" Lys33, Glu51, and Asp145
The 30mer peptide (numbers 67-96) shown in gold is a substrate for phosphorylation by the CDK2:cyclin A complex. It derives from an actual biological substrate in the protein cell division control protein 6 homolog, called CDC6-related protein. It is involved in a checkpoint control of the cell cycle that "checks" that DNA replication is completed before mitosis. It is discontinuous in the model since part of the bound peptide is intrinsically disordered and not observed in the crystal structure.
- The 1st fragment of the CDC6 peptide (67-73) contains the binding motif sequence S/T)PX(K/R) (the CDC6 substrate has the sequence 70Ser-Pro-Arg-Lys). Ser 70 is the target amino acid for phosphorylation by CDK2: cyclin A.
- The second fragment seen in the model (amino acids 85-96) contains another binding motif, RXL (in this peptide, RRL), which recruits cyclin A. This motif binds to the sequence MRAIL (210-214) in cyclin A.
Interestingly, this second binding site on cyclin A for its target protein is so far away from the active site of the CDK2:cyclin A. These kinds of interactions work to determine the specificity for CDKs and the binding cyclin partners.
CDKs 7, 9, 12, and 13 phosphorylate the RNA polymerase carboxy-terminal domain (CTD). The sequence of the CTD is unusual, composed of 52 heptad repeats in humans, with the consensus sequence YSPTSPS. Extracted from cells, CTD residues S2 and S5 are the most abundantly phosphorylated serine residues, while S7 is phosphorylated to a lesser extent. CDK7 has been shown to predominantly phosphorylate S5 and S7, CDK9 to have activity towards all three serines, and CDK12 and CDK13 to predominantly phosphorylate S2.
CDKs form complexes with target protein substrates and other proteins, which can serve as scaffolding anchors that bind both the CDK and the cyclin. Figure \(\PageIndex{9}\) shows an interactive iCn3D model of human CDK-activating kinase (CAK), a complex composed of cyclin-dependent kinase (CDK) 7, cyclin H, and the scaffolding protein MAT1 (6xbz).
.png?revision=1&size=bestfit&width=424&height=344)
The gray protein is CDK7, the cyan is Cyclin H, and the orange is MAT1. The purple again represents the C-helix of the CDK, and the red is the activation loop. The catalytic triad side chains in the CDK's active site are shown in CPK-colored sticks. Also shown is phospho-Ser in the activation loop.
The CDK activating kinase (CAK) shown above phosphorylates the target S/T in the activation loop (called the T-loop) in CDKs, activating the kinase. In addition, it regulates the initiation of transcription by phosphorylating the YSPTSPS repeats in the C-terminus of RNA polymerase II subunit RPB1. There are 15 consecutive repeats in the sequence, and others are dispersed in the C-terminal domain.
We already mentioned the motif RXL found in cyclin-binding proteins, which recruits them to cyclins (for example, through their interaction with MRAIL (210-214) in cyclin A. Likewise, short motifs in cyclins bind to proteins that increase or decrease CDK activity.
Several cyclin-encoded protein-binding sites or short peptide motifs have been structurally characterized. A well-characterized example is recycling the cyclin RXL recruitment site that is exploited to either enhance or inhibit CDK activity. Alternatively, short motifs encoded within the cyclin sequence can be used both to dock cyclins to substrates to enhance CDK activity and, alternatively, to localize them to CDK regulators, frequently resulting in a loss of CDK activity. Members of the p27KIP1/p21CIP1 cyclin-dependent kinase inhibitor (CKI) family share an RXL motif with RXL-containing substrates and compete with them for CDK–cyclin association. The INK (inhibitors of CDK) family of CKIs selectively inhibits CDK4 or CDK6 and, through an allosteric mechanism, disfavors CDK–cyclin binding. Their tandem ankyrin repeat structures exemplified by Cy CDK6–p19INK4d bind in the vicinity of the CDK hinge on the interface opposite to the surface remodeled upon cyclin association.
Modeling the Cell Cycle - Oscillations
Lastly, we will focus on mathematical models that show how the oscillatory behavior of CDK1/Cyclin B arises (remember that CDKs become active on binding to a cyclin). When CDK1 is activated, the cell is driven into mitosis. It is driven out of mitosis by activating the anaphase-promoting (APC) complex, which contains APC-Cdc20, an E3 ubiquityl ligase. Yeast Cdc20 is an activator protein that regulates the ubiquitin ligase activity of APC by binding at the right time in the cell cycle to B cyclins that contain a D box motif. This recruits the B cyclin:CDK1 to APC which ubiquitinates the cyclin B, leading to its degradation by the proteasome.
Figure \(\PageIndex{10}\) shows the levels of both cyclin B/CDK1 (red) and cyclin A/CDK2 (green) with time. Now in your mind, imagine another curve on the graph showing similar oscillations of activated APC, only frameshifted a bit in time so that the active APC trails that of active cyclin B/CDK1. When cyclin B/CDK1 is at its active peak, active APC is already beginning its rise.
Figure \(\PageIndex{10}\): The switch to sustained oscillations of cyclin A/Cdk2 (in green) and cyclin B/Cdk1 (in red) is shown following the overexpression of AP1. Gérard and Goldbeter, ibid.
The dissociation constant, KD, for CDK1 and cyclin B is about 28 nM, representing high-affinity binding. Cyclin B's ubiquitinylation and degradation allow for the freeing of CDK1 and inhibition of its activity. The oscillatory behavior in activity occurs only on the overexpression of yet another protein, AP1.
Let's explore in detail a model proposed by Ferrell et al that accounts for the 25 min oscillatory behavior of CDK1 in Xenopus (frog) eggs. The eggs are very large and perhaps because of their size have different constraints on their cell cle cycle. For example, cells can enter mitosis before the completion of DNA synthesis. The players that regulate their activity are shown below in Figure \(\PageIndex{11}\).
Figure \(\PageIndex{11}\): Proteins that help regulate the activity of CDK1 in Xenopus eggs.
- Cyclin: binds to and activates CDKs; active CDK1 drives cells into mitosis.
- APC: anaphase-promoting complex; active APC drives the cell out of mitosis with its E3 ubiquitin ligase activity, modifying cyclin and targeting it for proteolysis;
- Wee1: nuclear Ser/Thr kinase
- Cdc25: cell cycle division phosphatase that activates cyclin-CDK1.
Our goal in this discussion is to model the actual oscillatory behavior of CDK1 and show you how models are built and tested. When modeling enzyme inhibition data, it is important to fit the data to many models (reversible competitive, uncompetitive, mixed, and noncompetitive) to determine which model best fits the data. Typically, one starts with the simplest possible model and then advances to more complicated models until the data with the best statistical fit is found. Other examples, which are more relevant here, involve fitting binding and kinetic data using equations that give hyperbolic (for saturation binding and simple Michaelis-Menten kinetics) and sigmoidal fits (for cooperative binding and allosteric enzymes).
So, let's start with the simplest model that might lead to CDK1's oscillatory activity. In this section, the active form of CDK1 will be designated as CDK1*.
Model 1: One-step process with negative feedback - CDK1* inhibits its own activation (for example, by activating APC and hence ubiquitinylation of cyclin)
We saw in the Vcell models for the MAP kinase cascade that feedback phosphorylation of the first enzyme in the cascade (MAPKKK) by the last enzyme in the cascade, MAPKPP, leads to oscillations in enzyme activity. Could it explain the oscillations in the activity of CPK1 in Xenopus eggs? Let's make the following set of assumptions, which are all understandable from the material presented in previous chapters:
- CDK1* is inhibited by APC* (active APC), and to make it simpler, APC* can be expressed as a simple function of CDK1*, so there is just one species that varies in the flux equation;
- CDK1 activation occurs on rapid, high-affinity binding of cyclin, which is synthesized at a constant rate a1;
- The rate of CDK1 activation to produce CDK1* is given by mass action = rate activation - rate inactivation;
- APC is activated to APC "instantaneously" by CDK1*, so APC* is a very sensitive “cooperative” function of CDK1*, which can replace APC*. We will use the Hill equation for this "instantaneous" (or highly cooperative) effect, which gives a sharp, sensitive, cooperative rise in complex formation instead of a simple formation of a complex between CDK1* and APC. We explored this property of the Hill equation in Chapter 5.3.5 on Mathematical Analysis of Cooperative Binding.
Here is the Hill expression commonly used to fit the fractional saturation of a species empirically
\begin{equation}
Y=\frac{L^n}{K_D^n+L^n}
\end{equation}
It's a bit different than the Hill equation we saw for modeling the cooperative binding of O2 to hemoglobin (Chapter 5.3.5) since the KD term is also raised to the power n (which is not in the actual Hill equation). However, we did see that for oxygen binding to hemoglobin,
\begin{equation}
\mathrm{K}_{\mathrm{D}}=\mathrm{P}_{50}^{\mathrm{n}}
\end{equation}
Hence, the empirical expression used in fitting Model 1 is completely in accord with the Hill treatment of cooperativity. Again, we use the Hill equation when modeling binding and kinetic data that show significant sensitivity to conditions. It provides an additional parameter to aid in fitting the data and testing models.
Two different representations of a reaction diagram showing Model 1 are shown in Figure \(\PageIndex{12}\).
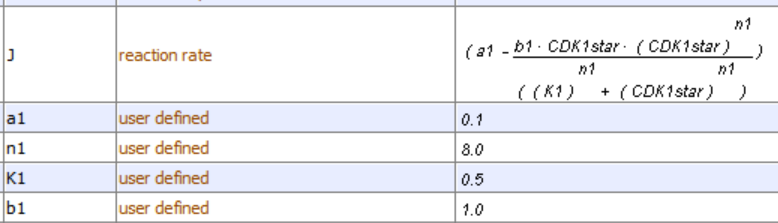
Figure \(\PageIndex{12}\): Two different representations of a reaction diagram for Model 1 - Activity of CDK1.
The representation on the right is from Vcell. The one on the left shows that CDK1 can inhibit itself.
Model 1 and its associated assumptions lead to the following differential equation that can be solved numerically in Vcell. All of the VCell outputs below were obtained from VCell models kindly provided by Leslie Loew.
\begin{equation}
v=\frac{d C D K 1^*}{d t}=a_1-b_1\left(C D K 1^*\right)\left(A P C^*\right)=a_1-b_1\left(C D K 1^*\right)\left(\frac{C D K 1^{* n 1}}{K_1^{n 1}+C D K 1^{* n 1}}\right)
\end{equation}
This rate equation has two terms (assumption 3). The first is the rate that CDK1* forms (a constant a1 defined by the rate of cyclin synthesis) and the rate at which it is degraded by APC*. The constant b1 in the second term can be thought of as a second-order rate constant for the interaction of CDK1* and APC*, a process that inactivates CDK1*.
The [APC*] in the middle equation is replaced with the Hill equation for APC's effective fractional saturation concentration (see assumption 4 of Model 1 described above) in the right-hand side.
\begin{equation}
\left(A P C^*\right)=\left(\frac{C D K 1^{* n 1}}{K_1^{n 1}+C D K 1^{* n 1}}\right)
\end{equation}
We will define the activity of the system as the rate at which CDK1* forms.
\begin{equation}
\text { Activity }=\frac{d C D K 1^*}{d t}
\end{equation}
Now let's see if changing the Hill coefficient n1 can cause oscillations in CDK1*.
Note that all the graphs plateau quickly, at which point the CDK1* activity is constant. The graph (gray) of the curve with the highest value of the Hill coefficient (n1=24) is linear and then abruptly plateaus. The slope of the velocity curve over the entire linear part of the n1=24 graph curve is 0.1, which is the value set for the rate of activation of CDK1. Then suddenly, at around 4 seconds, an "almost infinitely cooperative" shift to a constant formation rate occurs, arising from an abruptly reached rate of inactivation of CDK1* by the APC complex. These graphs do not show oscillations.
Now let's see the graph with no feedback inhibition, much as we did with the MAPK cascade in Chapter 12.4. The easiest way to do that is to set b1=0, the "second" order rate constant for the interaction of CDK1* and APC* in the model. The graph is shown in Figure \(\PageIndex{13}\).
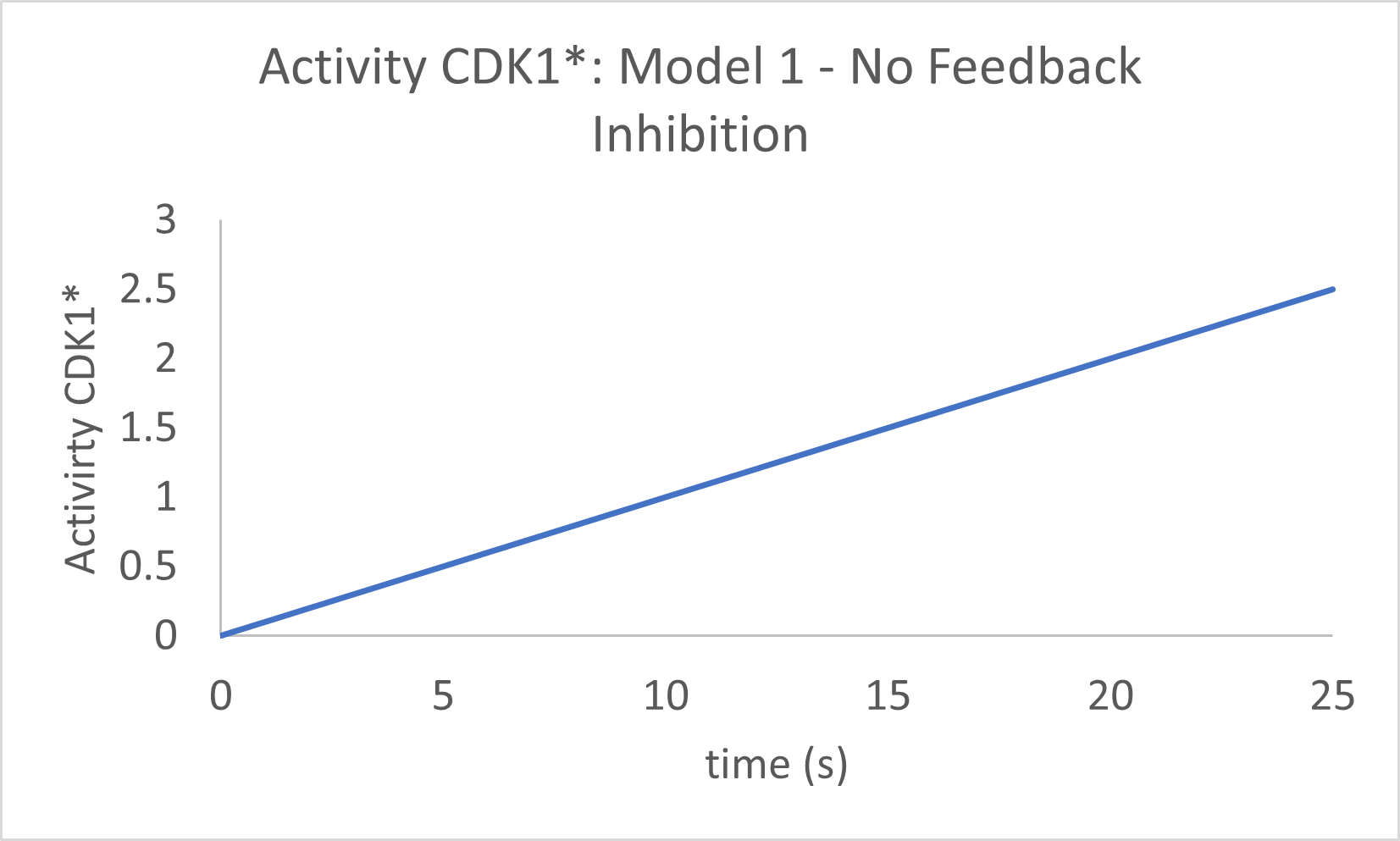
Figure \(\PageIndex{13}\): Activity of CDK1* in Model 1 in the absence of feedback inhibition.
The activity of CPK1* continually increases. When feedback inhibition is added, the curve "bends" to a plateau, but it does not start to decrease and shows no signs of oscillations. Time to move on to a more complex model!
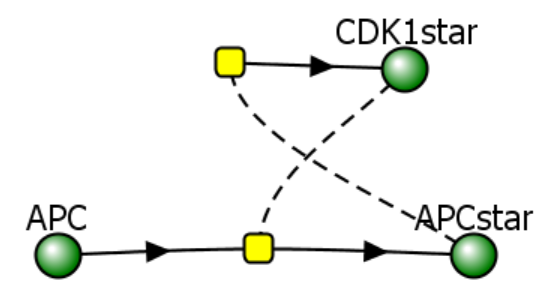
Model 2: Two-species model with activation and inhibition-
This model is more complicated and shows two species (CDK1 and APC) both of which are activated and inhibited. We need two mass action differential equations, one for each. Figure \(\PageIndex{14}\):
Figure \(\PageIndex{14}\): Two different representations of a reaction diagram of Model 2
The equation for dCDK1*/dt is the same as in Model 1, as is repeated below.
\begin{equation}
v=\frac{d C D K 1^*}{d t}=a_1-b_1\left(C D K 1^*\right)\left(A P C^*\right)=a_1-b_1\left(C D K 1^*\right)\left(\frac{C D K 1^{* n 1}}{K_1^{n 1}+C D K 1^{* n 1}}\right)
\end{equation}
Likewise, the equation for dAPC*/dt consists of two terms, one for its activation and one for its inhibition.
\begin{equation}
v=\frac{d A P C^*}{d t}=k_f A P C-k_r A P C^*
\end{equation}
Assume that kf, the rate constant of the activation of APC, is equal to a constant a2 times a Hill function of CDK1*, and kr, the rate constant for the inactivation of APC*, is simply b2. Then the equation becomes
\begin{equation}
v=\frac{d A P C^*}{d t}=a_2\left(\frac{C D K 1^{* n 2}}{K_2^{n 2}+C D K 1^{* n 2}}\right) A P C-b_2 A P C^*
\end{equation}
Let's look at the output graphs for the following initial condition:
- CDK1* = 0 uM
- APC = 1 uM
- APC* = 0 uM
Figure \(\PageIndex{15}\) shows static graphs of just CDK1* activity vs time for n1 values of 1, 4, 8, and 24 for Model 2.
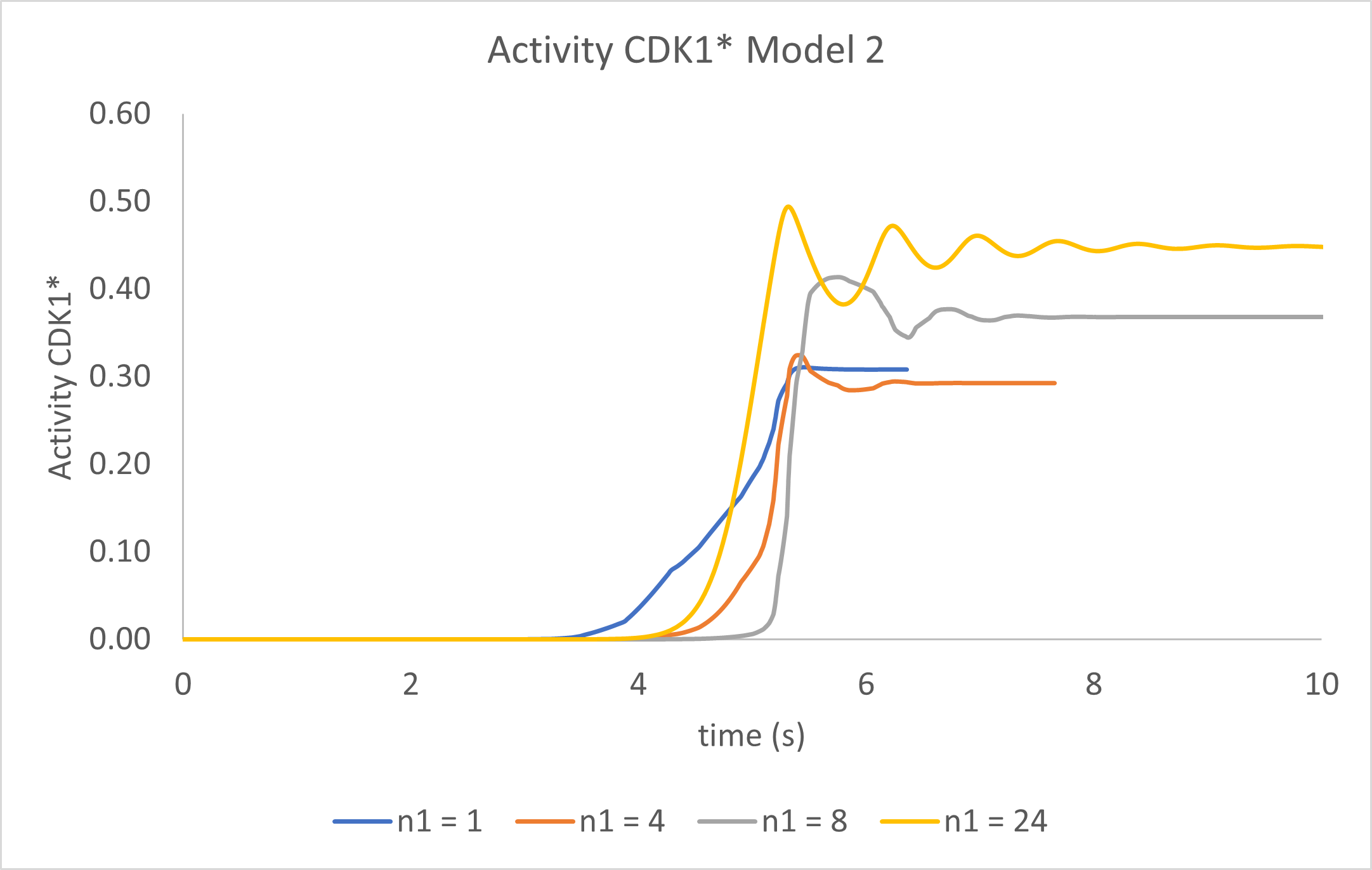
Figure \(\PageIndex{15}\): Graphs of just CDK1* activity vs time in Model 2 for n1 values of 1, 4, 8, and 24.
Wow! By simply adding an additional species to the model and a second differential equation for it, we see the first signs of oscillatory behavior in CDK1*'s activity. The output at higher n1 values is best described as damped oscillations. Now, let's try a final third model.
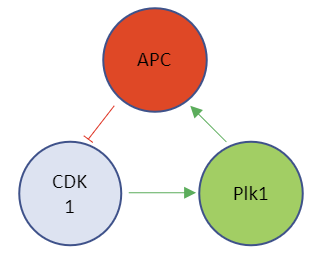
Model 3: Three-species model with activation and inhibition-
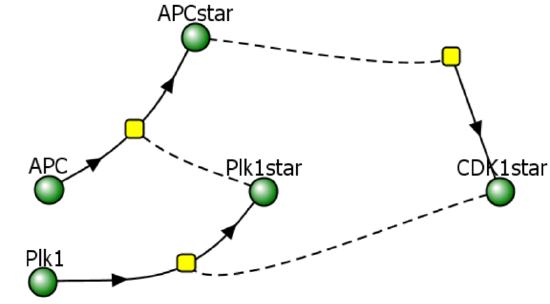
This model contains the enzyme Plk1 (Polo-like kinase 1, also called serine/threonine-protein kinase 10-A) and APC and CDK1. These three species are all activated and inhibited. Assume that CDK1 activates Plk1 and that it also helps activate APC. Following the arrows in the left part of the figure below shows that it acts as an "intermediary" between CDK1 and APC. Two different representations of a reaction diagram of Model 3 are shown in Figure \(\PageIndex{16}\).
Figure \(\PageIndex{16}\): Two different representations of a reaction diagram of Model 3
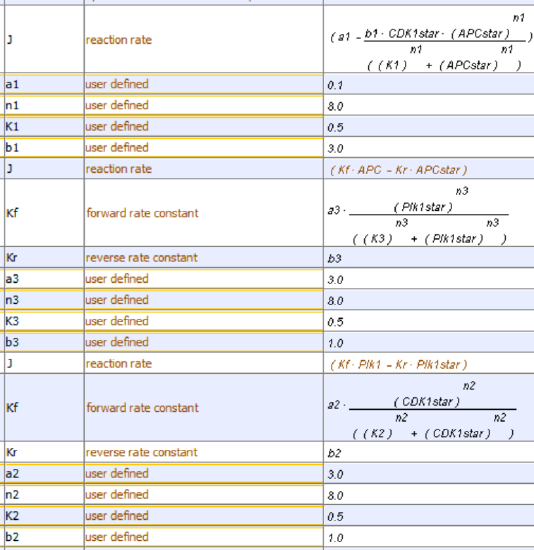
We have three species, so we need three differential equations, as shown below.
\begin{equation}
v=\frac{d C D K 1^*}{d t}=a_1-b_1\left(C D K 1^*\right)\left(\frac{A P C^{* n 1}}{K_1^{n 1}+A P C^{* n 1}}\right)
\end{equation}
\begin{equation}
v=\frac{d P l k 1^*}{d t}=a_2\left(1-P l k 1^*\right)\left(\frac{C D K 1^{* n 2}}{K_2^{n 2}+C D K 1^{* n 2}}-b_2 C D K 1^*\right.
\end{equation}
and
\begin{equation}
v=\frac{d A P C^*}{d t}=a_3\left(1-A P C^*\right)\left(\frac{P l k 1^{* n 3}}{K_3^{n 3}+P l k 1^{* n 3}}-b_3 A P C^*\right.
\end{equation}
The equation for activation of CDK1 is the same as in Models 1 and 2.
The equation for the activation of APC is similar to Model 2, with kf modeled as a Hill function of Plk1star, which activates APC
The equation for the activation of Plk1 is similar to Models 2 and 3, with kf modeled as a Hill function of CDK1star, which activates Plk1
Although you probably can't write these differential equations by yourself, hopefully, you can see that they make sense.
Figure \(\PageIndex{17}\) shows graphs of just CDK1* activity vs time for n1 values of 1, 4 and 8 for Model 3
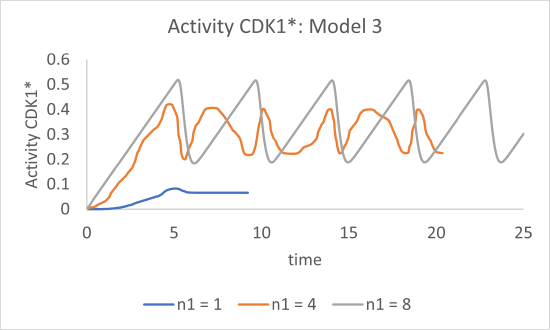
Figure \(\PageIndex{17}\): Graphs of CDK1* activity vs time for n1 values of 1, 4 and 8
Finally, we observe oscillatory behavior in the activity of CDK1*, but only for higher values of the Hill coefficient (n1 = 4 and 8).
Other models can produce oscillation, but this one seems perhaps most comprehensible to students who have studied mass action equations, Hill binding, and kinetic equations. Of course, the three-component system described in Model 3 is embedded in a large pathway of inputs and outputs, so other factors most likely affect the oscillatory behavior.
Summary
In this chapter, we explore the highly coordinated process of cell division in mammalian cells, which requires cells to progress through a series of ordered phases—G1, S, G2, and M—with the option to exit into quiescence (G0) or differentiate when necessary. Central to this regulation are cyclin-dependent kinases (CDKs), a family of serine/threonine protein kinases whose activities are tightly controlled by their binding to cyclins, as well as by CDK inhibitors (CKIs), phosphorylation/dephosphorylation events, ubiquitin-mediated degradation, and transcriptional regulation.
The chapter outlines how specific CDK-cyclin complexes function at different cell cycle stages:
- Early G1: CDK4/6 in complex with cyclin D responds to mitogenic signals by initiating phosphorylation of the retinoblastoma protein (pRb), which leads to the release of E2F transcription factors.
- Late G1/S Transition: CDK2, partnered first with cyclin E and later with cyclin A, completes pRb phosphorylation, triggering DNA replication in the S phase.
- G2 and Mitosis: CDK1 associates with cyclin A during G2 to prepare the cell for mitosis, while CDK1/cyclin B complex initiates mitosis. CDK1 activity is precisely controlled by activating phosphorylation (by the CDK-activating kinase, CAK) and inhibitory phosphorylation (by Wee1 and Myt1), with dephosphorylation by Cdc25 triggering entry into mitosis.
In addition to their roles in cell division, certain CDK complexes also participate in transcriptional regulation, DNA repair, and other cellular processes. The structural basis for CDK function is also discussed; key features include a bilobed structure, a flexible glycine-rich loop, the C-helix, and an activation loop that undergoes conformational changes upon cyclin binding and phosphorylation to enhance catalytic activity.
Finally, the chapter introduces mathematical models that illustrate how feedback loops and dynamic interactions among CDK1, cyclin B, the anaphase-promoting complex (APC), and other regulators generate the oscillatory behavior essential for proper cell cycle progression. Together, these mechanisms ensure that cell division is tightly controlled and that cells respond appropriately to internal and external signals, maintaining cellular homeostasis and organismal health.
This integrated view of cell cycle regulation provides a molecular framework for understanding not only normal cell proliferation but also the dysregulation that can lead to diseases such as cancer.