28.20: Signal Transduction - Pressure
- Page ID
- 80837
This chapter section is taken in entirety from Fang, XZ., Zhou, T., Xu, JQ. et al. Structure, kinetic properties, and biological function of mechanosensitive Piezo channels. Cell Biosci 11, 13 (2021). https://doi.org/10.1186/s13578-020-00522-z Creative Commons Attribution 4.0 International License, http://creativecommons.org/licenses/by/4.0/. We added iCn3D molecular models.
Introduction
Mechanotransduction, the process by which mechanical stimuli are converted into electrochemical signals, is essential for various biological processes, including neuronal cell development, pain sensation, and red blood cell volume regulation. As pivotal mechanosensors of in the mechanotransduction process, mechanosensitive (MS) ion channels have been found in organisms from bacteria to mammals. Extensive studies have revealed a variety of ion channels in eukaryotic cells that can sense various forms of mechanical forces (Table 1). These ion channels include transient receptor potential (TRP) channels and voltage-gated Na+, K+, and Ca2+ channels, whose dysfunction may be associated with human genetic diseases. Notably, the MS candidates identified in invertebrates either have no homologs (e.g., TRPN) or no functional conservation (e.g., DEG/ENaC/ASIC) in mammals. Furthermore, most MS candidates (the TRP channel in particular) are activated not only by mechanical stimuli but also by chemicals, temperature, osmolarity, and heat (> 27–34 °C). Defining the molecular details of MS cation channels in mammals is therefore of paramount importance to understand the mechanotransduction process and find potentially novel therapeutic strategies for mechanosensitivity disorders.
| Channel family | Channel isoforms | Ref. |
|---|---|---|
| TRP channels | TRPA1 | [6] |
| TRPC1 | [7] | |
| TRPC6 | [8] | |
| TRPV1 | [9] | |
| TRPV4 | [10] | |
| TRPM4 | [11] | |
| TRPM7 | [12] | |
| TRPN | [13] | |
| TRPP2 | [14] | |
| K + channels | Shaker (Kv1.1) | [15] |
| Ca2+-activated K+ (BK) | [16] | |
| TREK1/2 | [17] | |
| TRAAK | [18] | |
| HCN2 | [19] | |
| Na+ channels | Nav1.5 | [20] |
| Ca2+ channels | L-type | [21] |
| N-type | [22] | |
| T-type | [23] | |
| Cl− channels | CFTR | [24] |
| OSCA protein family | ScCSC1, HsCSC1 | [25] |
| DEG/ENaC superfamily | C.elegans MEC (MEC-4, MEC-10) | [26] |
| ASIC | [27] | |
| Other channels | TMC1/2 | [28] |
In 2010, Coste et al. revealed a novel family of mechanically activated (MA) cation channels in eukaryotes consisting of Piezo1 and Piezo2 channels, which have been proposed as the long-sought-after MS ion channels in mammals. The Piezo1 channel is present in nonsensory tissues, with particularly high expression in the lung, bladder, and skin; by contrast, the Piezo2 channel is predominantly present in sensory tissues, such as dorsal root ganglia (DRG) sensory neurons and Merkel cells. Since their discovery, tremendous effort has been made to reveal the structures and biological functions of Piezo 1 and 2. The partial molecular structure of a Piezo channel was determined by cryo-electron microscopy (cryo-EM). Furthermore, Piezo channels have been linked to various pathological and physiological processes, including erythrocyte volume regulation, cell division, and innate immunity. Moreover, Piezo channel mutations are associated with multiple hereditary human diseases, such as autosomal recessive congenital lymphatic dysplasia, hereditary xerocytosis (a rare, autosomal dominant congenital hemolytic anemia characterized by macrocytic stomatocytosis, and decreased red cell osmotic fragility due to a defect in cation permeability), and an autosomal recessive syndrome of muscular atrophy with perinatal respiratory distress. Considerable progress has been made toward characterizing the structural features, physiological significance, and biophysical properties of Piezo proteins. Given the importance of Piezo channels in understanding mechanotransduction processes, this review focuses on their structural details, kinetic properties, and potential functions as mechanosensors. We also briefly review the hereditary diseases caused by mutations in the Piezo genes, which is key to understanding their functions.
Structure of Piezo channels
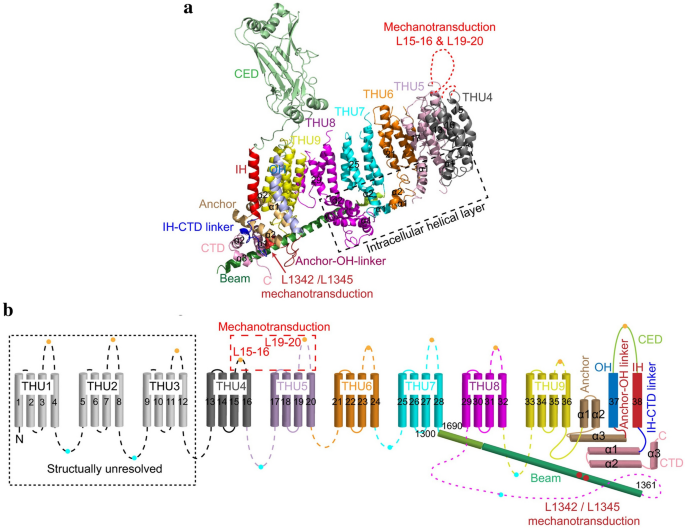
Piezo proteins have an uncommonly large predicted size of approximately 2500 amino acids and encompass numerous transmembrane (TM) regions. Subsequent research has revealed that the mouse Piezo1 (mPiezo1) channel is an evolutionarily conserved pore-forming ion channel directly gated by membrane stretch. Several published cryo-EM studies have revealed that mPiezo1 exhibits a three-bladed, propeller-shaped homotrimeric structure that includes a central cap, three peripheral blade-like structures on the extracellular side, three long beams on the intracellular side that bridge the blades to the cap, and a TM region between these features (Fig. 1).
Figure 1: Cryo-EM structure of the mPiezo1 channel (adapted from Zhao et al.). a Multiple views of the sharpened map of the trimeric channel with the major domains labeled, with the three subunits colored red, green, and blue. b Cartoon model in which the three subunits are colored red, green, and blue. In the middle panel, the front subunit has been omitted to provide a better view of the curvature of the TMs
Structure of the Piezo1 channel
Unprecedented 38-TM topology
Piezo channels are predicted to possess an unusually large number of TM regions, ranging from 10 to 40. Zhao et al. recently produced high-resolution structures of mouse Piezo1 (mPiezo1), revealing a unique 38-TM topology in each subunit (Fig. 2a, b). The two TM regions (TM37 and TM38) closest to the center of the protein are designated as the inner helix (IH) and outer helix (OH), respectively, and enclose the transmembrane pore of the central pore module. The other 36 TM regions (TM1-36) form a curved blade-like structure with nine repetitive folds containing 4 TM regions each, named transmembrane helical units (THUs)
Figure 2: A 38-TM topology model and key functional sites in mPiezo1(adapted from Zhao et al.). a A model showing one subunit with individual THUs and featured structural components. Residues L1342 and L1345 in the beam are indicated by red spheres. b A 38-TM topology model color-coded to match the cartoon model in A
Figure \(\PageIndex{a}\) shows an interactive iCn3D model of the mouse mechanosensitive Piezo1 channel (5Z10) (long load time)
.png?revision=1&size=bestfit&width=412&height=280)
Central cap
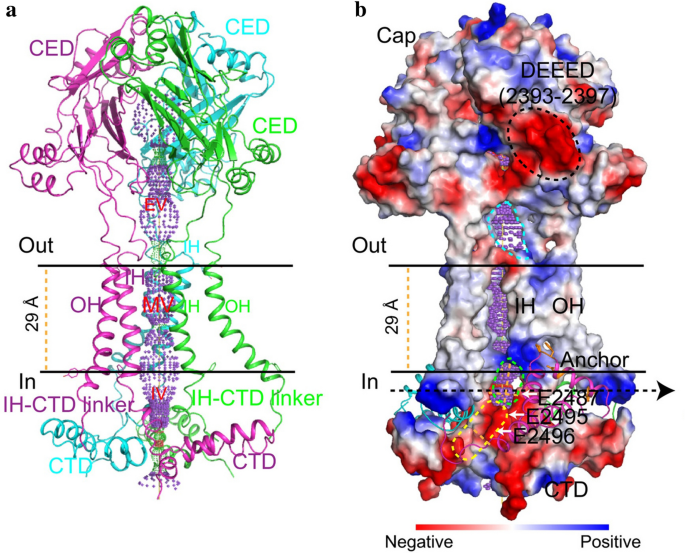
Kamajaya and colleagues [48] employed topological prediction modeling and found that residues 2210 to 2457 in Piezo1 form an extracellular loop following the last TM region from the C-terminus, defined as the C-terminal extracellular domain (CED) (Fig. 1). The deletion of residues 2218 to 2453 from the Piezo1 protein abolished expression of the central cap, suggesting that this region trimerizes to form the central cap (Figs. 1 and 3). Further analysis revealed that the central cap consists of the CED in the form of a trimeric complex that encloses an extracellular vestibule (EV) with openings (Fig. 3).
Anchor
A hairpin structure, referred to as the anchor, connects the OH-IH pair to the C-terminal domain (CTD) plane, which moves the OH-CED-IH-containing region of one subunit into the neighboring subunit in a clockwise direction (Figs. 1 and 2). The anchor is made up of three helices (α1, α2, and α3). Helices α1 and α2 were found to organize into an inverted V-shaped structure, which maintains the integrity of the ion-conducting pore (Fig. 2b). In parallel with the membrane plane, the long α3 helix links to the OH via a lysine-rich anchor-OH linker that interacts with the polar residue-rich α2–3 turn in the anchor and the glutamate-rich region of the CTD. A few mutations in Piezo1 at locations including KKKK (2182-K2185), T2143, T2142 (T2127 in human Piezo1), R2514, E2523, and E2522, which are located in α3 in the anchor, have been reported to cause severe disease. Additionally, SERCA2, a Piezo-interacting protein, suppresses Piezo1 by acting on the anchor-OH linker. These findings support the structural and functional importance of the anchor region.
The long intracellular beam
On the intracellular surface, Piezo1 contains three beam-like structures 90 nm in length that are organized at a 30° angle relative to the membrane plane (Figs. 1 and 2). Residues H1300-S1362 form the beam structure. The large intracellular THU7-8 loop, which contains approximately 390 residues, might provide the beam with the structural basis for mechanical transmission. Functionally, the three long intracellular beams not only support the whole TM skeleton but also physically bridge the distal THUs to the central ion-conducting pore. When residues 1280 to 1360 (which form this beam structure) were deleted, the resulting mutant protein was absent, suggesting the structural importance of the beam.
Highly curved blades
The nine peripheral THUs in each subunit form blade-like structures, with each blade twisted clockwise (Fig. 1b). The proximal TM25–TM36 and peripheral TM13-24 interact at a 100° angle, as viewed from 90º relative to the plasma membrane plane, and a 140° angle, as viewed from a line parallel to the plasma membrane plane. Another important feature of the blades is the L-shaped helical structures formed by TM13, TM17, TM21, TM25, and TM29. Both identifiable structural features appear to be ideal not only for mechanosensation but also for the induction of local membrane curvature. Intriguingly, the peripheral TM13-24 appears to be within a highly curved membrane plane, indicating that the Piezo1 channel can curve the membrane in which it resides. This is consistent with past studies implying that Piezo1 ion channels can be regulated by cellular membrane curvature and tension.
The ion-conducting pathway
As pore-forming ion channels, Piezo proteins contain a trimeric ion-conducting channel made up of residues 2,189 to 2,547, which contain the last two TMs (Fig. 3). The continuous central channel consists of three parts, an EV within the cap region, a transmembrane vestibule (MV) within the membrane, and an intracellular vestibule (IV) underneath the membrane. Both the EV and IV possess an opening that connects to the MVs, which are positioned above and below the membrane. Importantly, DEEED (2393–2397), a patch of negatively charged residues residing in the opening of the extracellular “cap” structure consisting of the CED, is required to ensure efficient ion conduction and determine the selection of cations over anions. Additionally, two critical acidic residues, E2495 and E2496, located at the CTD-constituted IV, may be responsible for divalent calcium ion selectivity, unitary conductance, and pore blockage.
Structure of the Piezo2 channel
Similar to Piezo1 channels, Piezo2 channels are large membrane proteins consisting of over 2,800 residues. However, the Piezo2 channel and Piezo1 channel share approximately only 42% sequence homology. Recent studies have shown that the overall structure of the Piezo2 channel is very similar to that of Piezo1 in that it forms a three-bladed, propeller-like homotrimeric structure comprising a central ion-conducting pore module and three peripheral blades with 38 TMs.
Figure \(\PageIndex{b}\) shows an interactive iCn3D model of the mammalian tactile channel PIEZO2 (6KG7) (long load time)
2.png?revision=1&size=bestfit&width=456&height=321)
In the Piezo2 channel, the charged residues at the interface between the beam and the CTD are required to ensure the normal mechanosensitivity of the channel. Moreover, single-channel recordings indicated that a previously unrecognized intrinsically disordered domain adjacent to the beam acts as a cytosolic plug that limits ion permeation, possibly by clogging the inner vestibule in both Piezo1 and Piezo2. Furthermore, by structurally comparing the Piezo1 and Piezo2 channels, Wang et al. found that the Piezo2 channel has additional constriction sites at L2743, F2754, and E2757 that might serve as a transmembrane gate controlled by the cap domain.
Lever-like mechanotransduction mechanism
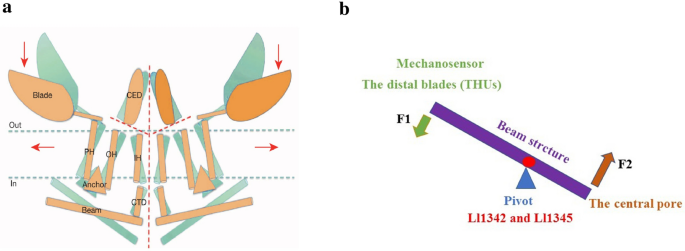
Based on the unique topological features of the mPiezo1 channel, a lever-like mechanotransduction mechanism to explain its extraordinary mechanosensitivity was proposed (Fig. 4). In the mPiezo1 channel, the curved blades composed of THUs can act as a mechanosensor, while the beam structure, with the residues Ll1342 and Ll1345 acting as a pivot, can act as a lever-like apparatus. Coupling the distal blades and the central pore through the lever-like apparatus converts mechanical force into a force used for cation conduction.
Adapted from Ge et al. b A lever-like mechano-gating model in Piezo1. The blue and red dashed arrows indicate input and output forces, respectively
Figure 4: Model of the lever-like mechanotransduction model. The curved blades can act as a mechanosensor, while the beam structure, with residues Ll1342 and Ll1345 acting as a pivot, can act as a lever-like apparatus. Coupling of the distal blades and the central pore through the lever-like apparatus converts mechanical force into cation conduction. a Proposed model of the force-induced gating of Piezo channels. The blue and orange models represent the channel in its closed and open states, respectively. Red dashed lines indicate possible ion-conduction pathways.
Because the pivot of the lever is positioned closer to the central pore than to the distal blades, the input force is effectively amplified through the lever-like apparatus. Additionally, a large conformational change in the distal blades is converted into a relatively slight opening of the central pore, allowing cation-selective permeation.
Kinetics properties of Piezo channels
Activation mechanisms of Piezo channels
Normal Piezo channel kinetics can be modeled with three states: open, closed, and inactivated; these states have emerged, collectively, as an important mechanism in the Piezo channel function. Studies have proposed that the Piezo1 channel is gated directly by bilayer tension that can be modified by cytoskeletal proteins and linkages to the extracellular matrix (ECM). For example, in overhydrated red blood cells (RBCs), Piezo1-mediated Ca2+ influx activates K+ efflux through the Gardos channel (KCa3.1), which in turn leads to water loss and RBC dehydration.
Piezo1 and Piezo2 channels not only exhibit a three-bladed, propeller-shaped trimeric architecture but also can locally deform lipid membranes into a dome-like shape. In addition, changes in the projection area of Piezo channels from closed to open are essential for their mechanosensitivity; this was investigated by calculating the available free energy. Based on these findings, the membrane dome mechanism was proposed and experimentally proved to explain the activation mechanisms of Piezo channels (Fig. 5). Essentially, the dome shape created by Piezo channels in their closed conformation acts as a potential energy source for MS gating. Under tension, lateral membrane tension flattens the Piezo dome, which increases the energy of the membrane-channel system in proportion to the expansion of the projected area of the dome. Piezo channels then open due to the relative energy difference. This mechanism can account for the highly sensitive mechanical gating of Piezo channels with a cation-selective pore. Although the membrane dome mechanism explains the exquisite mechanosensitivity of Piezo channels, it does not consider the shape of the surrounding membrane. Haselwandter et al. [57] proposed the membrane footprint hypothesis, which states that the Piezo1 channel deforms the shape of the membrane outside the perimeter of the channel such that it exhibits a curved membrane footprint, which amplifies the sensitivity of Piezo1 to changes in the membrane tension. Nevertheless, further experiments are needed to test and refine these hypotheses.

Inactivation kinetics of Piezo channels
Various types of mechanical stimulation trigger Piezo channel activation and sequentially elicit an MA current with rapid decay, even in the presence of continued stimulation, due to rapid inactivation. Coste et al. first described detailed information about the voltage-dependent inactivation kinetics of Piezo channels, characterized as fast at rather negative membrane potentials and slow at rather positive membrane potentials. Additionally, Piezo1 channel inactivation is relatively slow compared with Piezo2 channel inactivation. Several point mutations in Piezo channels have been reported to slow down the inactivation process, which produces larger cation fluxes and results in various human diseases. Given its demonstrated key role in normal channel function, we next review what is known about the inactivation kinetics of Piezo channels with a focus on the inactivation mechanism.
The available information regarding the structures (residues/domains) and human disease-related point mutations have helped to clarify the mechanisms of ion channel inactivation. Currently, six gain-of-function mutations associated with dehydrated hereditary xerocytosis (DHS) have been found to slow the inactivation rate of Piezo channels (Table 2), most of which are clustered at the central core region of the Piezo channel structure. This implies that the pore region, which contains an OH, an IH, an extracellular cap domain, and an intracellular CTD, determines the kinetics of inactivation. Further detailed links between structural domains and inactivation kinetics have been investigated. Wu et al. identified that the distinct inactivation kinetics of Piezo1 and Piezo2 channels and characteristic voltage-dependent inactivation appears to be determined by the C-terminal extracellular domains (cap domain). Two potential inactivation gates within the IH and CTD have been thought to be sufficient for the normal inactivation of the Piezo1 and Piezo2 channels. Recently, three small subdomains within the extracellular cap were shown to individually confer Piezo channel inactivation. These results support the idea that the ion-conducting pore region of Piezo channels is essential for their inactivation properties.
Table 2 Mutations in Piezo1 and Piezo2 Associated with Human Diseases: Full-size table
Interestingly, a slowly inactivating MS current in mouse embryonic stem cells (mESs) has been described that is dependent on the Piezo1 channel. However, heterologous expression of Piezo1 cDNA from mES cells displays fast inactivation kinetics, indicating that additional regulatory mechanisms other than the amino acid sequence determine the slow kinetics of the Piezo1 channel in mES cells [70]. Recently, sphingomyelinase activity has been revealed to be a crucial determinant of Piezo1 inactivation. Various modulators, such as pH, temperature, divalent ion concentrations, alternative splicing, osmotic swelling, membrane lipid composition, co-expression of other membrane proteins, and G-protein-coupled pathways have also been reported to regulate the Piezo channel kinetics; however, we still know very little about the relationships among these factors and pivotal structural domains.
Pharmacological modulators of Piezo channels
Despite the relatively recent discovery of Piezos, there has been progress regarding small-molecule modulators of Piezo1. Piezo1 chemical activators, including Yoda1 and Jedi1/2, were able to open Piezo1 ion channels in the absence of mechanical stimulation. Jedi1/2, a novel hydrophilic Piezo1 chemical activator, acts through the peripheral blades and utilizes a peripheral lever-like apparatus consisting of the blades and a beam to gate the central ion-conducting pore whereas Yoda1 acts as a molecular wedge, facilitating force-induced conformational changes, effectively lowering the channel’s mechanical threshold for activation However, the reason why Yoda1 does not efficiently activate the Piezo2 channel is unclear. Specific inhibitors of Piezo1 are in their infancy. As nonspecific inhibitors of the ion pore in stretch-activated ion channels, gadolinium, and ruthenium red have also been shown to block mouse Piezo1 channels with IC50 values of approximately 5 mM. The commonly used toxin inhibitor of mechanosensitive channels, GsMTx4, was also found to inhibit the Piezo1channel, but it might not bind Piezo1 directly, rather acting via modulating local membrane tension near the channel. Dooku1, an analog of Yoda1 without a stimulatory effect, antagonizes Yoda1-evoked activation of Piezo1 and aortic relaxation.
The function of Piezo channels
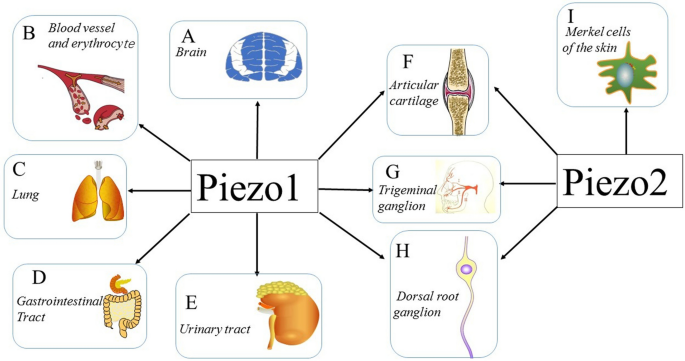
Piezo channels are expressed in a wide range of mechanically sensitive cells and allow Ca2+ influx in response to different types of external forces, such as fluid flow, pulling, and ultrasonic forces. The biological function of Piezo channels was recently investigated in a number of studies (Fig. 6). The results of these studies verified the pivotal roles of Piezo channels in mechanotransduction under physiological and pathophysiological conditions. Here, we focus on reviewing the biological function of Piezo channels in several different types of MS tissues and cells.
Figure 6: Expression and function of Piezo channels Multiple tissues and cells express Piezo channels, and each of those shown is discussed in this review. a–e demonstrate the vital role of the Piezo1 channel in the CNS, blood vessels, erythrocytes, lungs, gastrointestinal tract, and urinary tract. f–h illustrate the expression of both the Piezo1 channel and Piezo2 channel in articular cartilage, trigeminal ganglia, and dorsal root ganglia. i shows that the Piezo2 channel is expressed in Merkel cells, which are involved in sensing light touch
Consult the original article for details on the roles of Piezo channels on the systems above.