14.4: Concentration Control and Elasticity Coefficients
- Page ID
- 68905
Concentration Control Coefficients
The Concentration control coefficient (\(C_{E_{i}}^{S}\)), a global property of the system, gives the relative fractional change in metabolite concentration \(S_j (dS_j/S_j)\), where \(S_j\) is the concentration of any metabolite in the system and as such is a system variable, with fractional change in concentration or activity of enzyme \(E_i (du_i/u_i)\). Similar equations as section 15.3 apply.
\begin{equation}
C_{E_i}^S=\frac{\frac{\partial S_i}{S_i}}{\frac{\partial u_i}{u_i}}=\frac{\partial \ln S_i}{\partial \ln u_i}=\frac{\partial S_i}{\partial u_i} \frac{u_i}{S_i}
\end{equation}
It can be shown that the sum of all of the individual \(C^S_{E_i} = 0\) (another summation theorem), is not 1 as in the case of flux control coefficients. This again would make sense in the steady state. Flux coefficients usually vary from 0 to 1, but concentration coefficients can vary from negative to positive and small to large.
\begin{equation}
\sum_{i=1}^n C_{E i}^S=0
\end{equation}
The concentration control coefficients can have large values as seen in a simple example. For a given enzyme, at low [S], for example, when [S] << Km,
\begin{equation}
v=\frac{V_m S}{K_M+S}=\frac{V_m S}{K_M}=\frac{k_{c a t} E_{t o t} S}{K_M} \text { when } S \ll K m
\end{equation}
If the enzyme had only 0.1x of its normal activity (due to a mutation for example), then to maintain constant flux, the [S] would have to increase 10-fold.
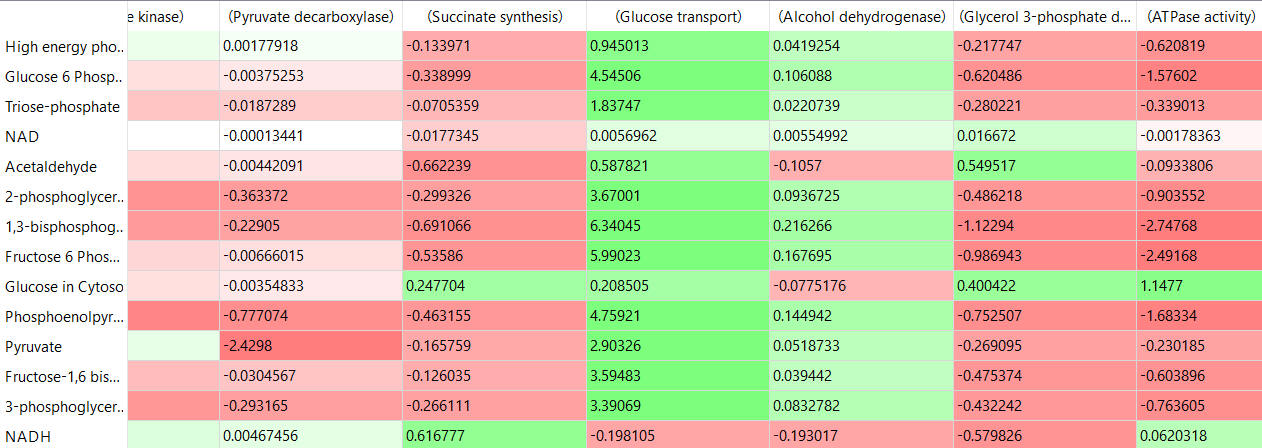
The tables below show the CSEi values for incremental changes in the substrate (1%).
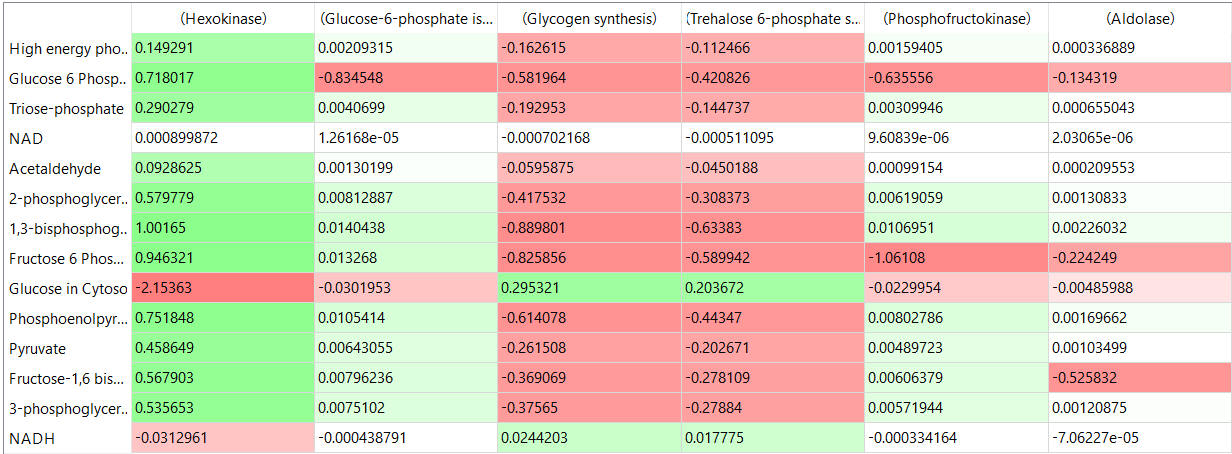
table, part 2:
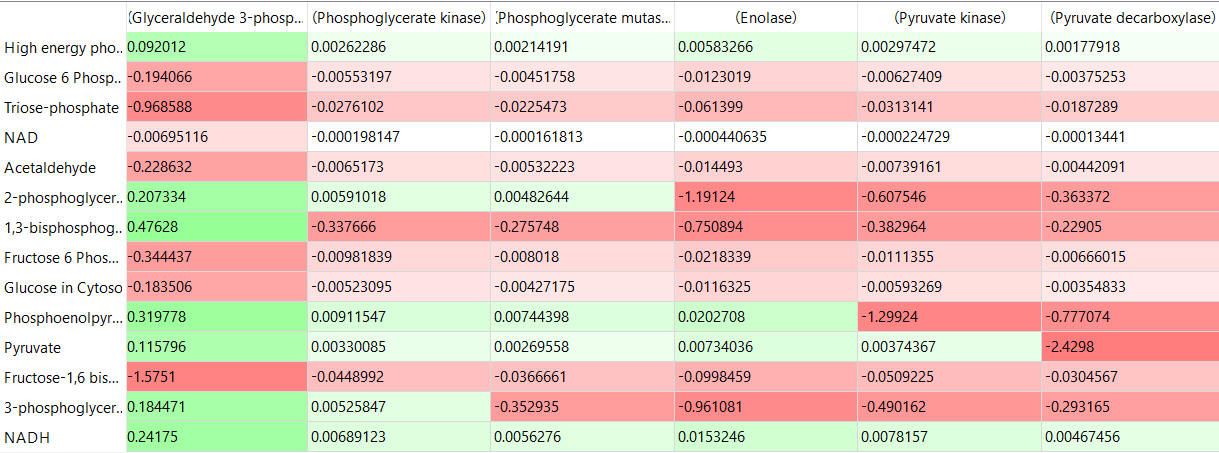
table, part 3
Elasticity Coefficient
The Elasticity Coefficient, in contrast to the flux and concentration control coefficients, which are properties of the system, is a local property and can be measured using isolated enzymes and substrates. It makes sense that some kinetic property of the isolated enzyme would affect its propensity to affect system flux. The elasticity coefficient gives a measure of how much a substrate \(S\) (or other substance) can change the reaction rate (\(v\)) of an isolated enzyme. (Note we use \(v\) and not flux \(J\), which describes a system property.) Hence
\begin{equation}
\varepsilon_S^v=\frac{\frac{\partial v}{v}}{\frac{\partial S}{S}}=\frac{\partial \ln v}{\partial \ln S}=\frac{\partial v}{\partial S} \frac{S}{v}
\end{equation}
The term elasticity is also used in economics and is especially useful in times when inflation is high. If the price of your favorite product, such as a Starbucks Latte coffee goes up, consumers might either buy them less frequently or buy another cheaper latte from a competitor. If a small rise for a Starbucks latte leads to a large drop in demand, the Starbucks latte is characterized by a high elasticity. If however, people don't change their latte buying behavior when there is a big price rise on the latte, the product is said to be inelastic. If you are the CEO of a company, it is good to know the elasticity of your products to maximize your profits.
Hence the elasticity coefficient can be determined using basic enzyme kinetics of the isolated enzyme. Note that the coefficient at each \(S\) concentration is the slope of the v vs S curve multiplied by the \(S/v\) at that tangent point. The elasticity coefficient must be evaluated at the same concentration of enzyme and substrate as found in vivo in the steady state. Velocity (\(v\)), not flux, is used. In the above case, \(S\) is the substrate, but it could be a product or modifier. There is a different elasticity coefficient for each parameter. There is no summation theory for elasticities. Values can be positive for species that increase the velocity or negative for those that decrease it. Hence there can be multiple elasticities.
The tables below show the elasticity coefficients (relative or scaled) for yeast glycolysis determined using COPASI.
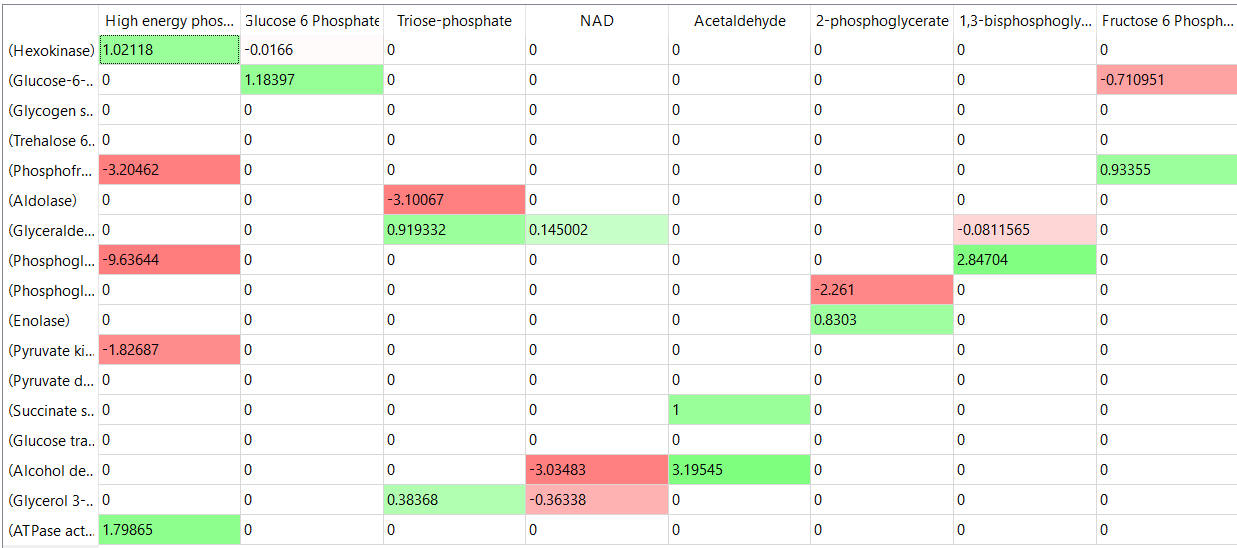
table, part 2
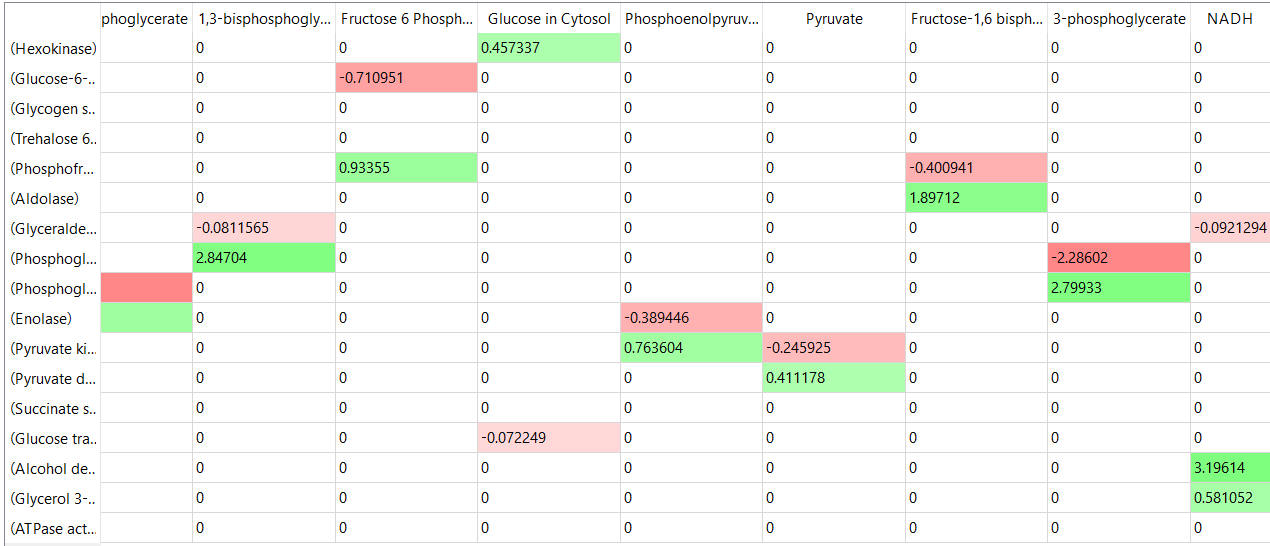
Things to note:
- the columns show substrates not enzymes;
- green cells (with positive elasticities) are generally substrates for their target enzymes (for example, glucose-6-phosphate for phosphoglucomutase);
Other "generic" sensitivities can be determined as well. For example, incremental changes in \(K_m\), \(V_m\), or \(K_{ix}\) for specific enzymes could affect fluxes in a pathway.
A link between system control coefficients and local coefficients:
You would think that there should be some relationship between a system variable such as the flux control coefficient and a local variable such as the elasticity coefficient. There is and it is expressed by the Connectivity Theorem (below). If a substrate \(S\) is acted upon by many different enzymes (\(i …… n\)), which is very likely, especially for branch points in metabolic pathways, then it can be shown that
\begin{equation}
\sum_{i=1}^n C_{v i}^J e_s^{v i}=0
\end{equation}



