13.2B: Cytolysis by the Membrane Attack Complex (MAC)
- Page ID
- 3314
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Discuss how antibodies defend the body by way of MAC cytolysis. (Include what classes or isotypes of immunoglobulins are involved, the role of the Fab portion of the antibody, the role, if any, of the Fc portion of the antibody, and the role of any complement proteins, if any, involved.)
- State specifically how MAC cytolysis protects against the following:
- Gram-negative bacteria
- human cells recognized as nonself
- enveloped viruses
- Describe one way Gram-negative bacteria may resist cytolysis.
The process starts with the antibody isotypes IgG or IgM being made against epitopes on membranes. The Fab portion of IgG or IgM reacts with the epitopes on the membrane and the Fc portion of the antibody then activates the classical complement pathway. C5b6789n (the membrane attack complex or MAC) then puts holes in the membrane. (Remember that MAC is also produced during the alternative complement pathway and the lectin pathway as was discussed in Unit 5.)
a. In the case of bacteria, MAC can put holes in the outer membrane and possibly the cytoplasmic membrane of the Gram-negative cell wall (Figure \(\PageIndex{1}\) left) causing lysis ( Figure \(\PageIndex{1}\) right).
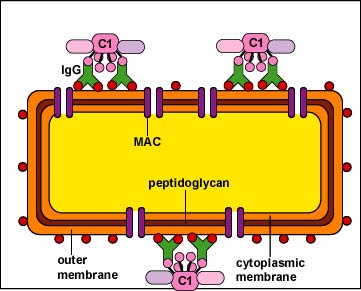
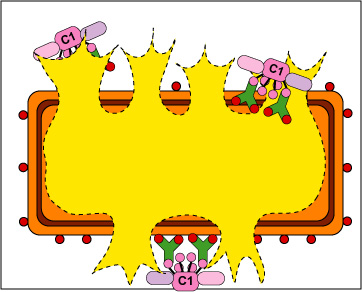
b. With enveloped viruses, the MAC can damage the viral envelope (Figure \(\PageIndex{12}\).3.2).
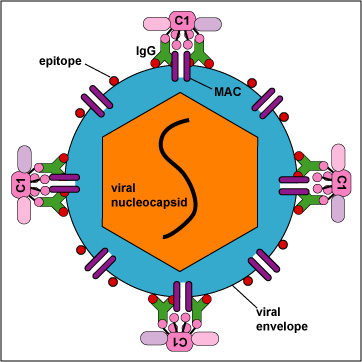
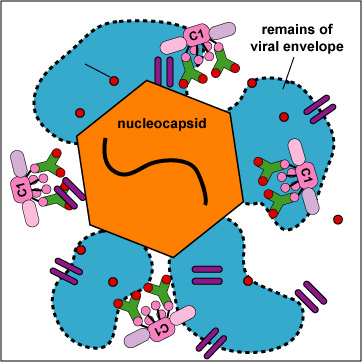
c. In the case of human cells recognized as nonself- virus-infected cells, transplanted cells, transfused cells, cancer cells - the MAC causes direct cell lysis (see Figure \(\PageIndex{5}\) and Figure \(\PageIndex{6}\)).
Concept Map for Ways in which Antibodies Protect the Body
However, as learned in Unit 3, some bacteria by means of the activities described below are more resistant to MAC lysis.
- The LPS of the cell wall is the principle target for complement in Gram-negative bacteria by activating the alternative complement pathway and serving as a binding site for C3b as well as the site for formation of MAC. Some Gram-negative bacteria attach sialic acid to the LPS O antigen (see Figure \(\PageIndex{7}\)) and this prevents the formation of the complement enzyme C3 convertase that is needed for the eventual formation of all the beneficial complement proteins such as C3b, C5a, and MAC. Blood-invasive strains of Neisseria gonorrhoeae , as well as Bordetella pertussis and Hemophilus influenzae are examples of Gram-negative bacteria that are able to alter their LPS in this manner.
- Some Gram-negative bacteria, such as Salmonella , lengthen the LPS O antigen side chain (see Figure \(\PageIndex{7}\)) and this prevents the formation of MAC. Neisseria meningitidis and Group B Streptococcus , on the other hand, produces capsular polysaccharides composed of sialic acid and as mentioned above, sialic acid prevents MAC lysis.
- An outer membrane molecule of Neisseria gonorrhoeae called Protein II binds factor H of the complement pathway and this leads to the degradation of the opsonin C3b by factor I and the formation of C3 convertase. Without C3 convertase, no MAC is produced.
Summary
The Fab portion of IgG or IgM reacts with the epitopes on the membrane and the Fc portion of the antibody then activates the classical complement pathway. C5b6789n (the membrane attack complex or MAC) then puts holes in the membrane. In the case of bacteria, MAC can put holes in the outer membrane and possibly the cytoplasmic membrane of the Gram-negative cell wall causing lysis. In the case of enveloped viruses, MAC can damage the viral envelope. In the case of human cells recognized as nonself - virus-infected cells, transplanted cells, transfused cells, cancer cells- the MAC causes direct cell lysis.
Questions
Study the material in this section and then write out the answers to these questions. Do not just click on the answers and write them out. This will not test your understanding of this tutorial.
- Discuss how antibodies defend the body by way of MAC cytolysis. (Include what classes or isotypes of immunoglobulins are involved, the role of the Fab portion of the antibody, the role, if any, of the Fc portion of the antibody, and the role of any complement proteins, if any, involved.) (ans)
- Some Gram-negative bacteria attach sialic acid to the LPS O antigen of their outer membrane. Briefly describe how this may protect that Gram-negative bacterium from MAC cytolysis. (ans)
- How does MAC affect viruses? (ans)
- Multiple Choice (ans)


