6.5: Enzymes
- Page ID
- 1854
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Skills to Develop
- Describe the role of enzymes in metabolic pathways
- Explain how enzymes function as molecular catalysts
- Discuss enzyme regulation by various factors
A substance that helps a chemical reaction to occur is a catalyst, and the special molecules that catalyze biochemical reactions are called enzymes. Almost all enzymes are proteins, made up of chains of amino acids, and they perform the critical task of lowering the activation energies of chemical reactions inside the cell. Enzymes do this by binding to the reactant molecules, and holding them in such a way as to make the chemical bond-breaking and bond-forming processes take place more readily. It is important to remember that enzymes don’t change the ∆G of a reaction. In other words, they don’t change whether a reaction is exergonic (spontaneous) or endergonic. This is because they don’t change the free energy of the reactants or products. They only reduce the activation energy required to reach the transition state (Figure \(\PageIndex{1}\)).

Enzyme Active Site and Substrate Specificity
The chemical reactants to which an enzyme binds are the enzyme’s substrates. There may be one or more substrates, depending on the particular chemical reaction. In some reactions, a single-reactant substrate is broken down into multiple products. In others, two substrates may come together to create one larger molecule. Two reactants might also enter a reaction, both become modified, and leave the reaction as two products. The location within the enzyme where the substrate binds is called the enzyme’s active site. The active site is where the “action” happens, so to speak. Since enzymes are proteins, there is a unique combination of amino acid residues (also called side chains, or R groups) within the active site. Each residue is characterized by different properties. Residues can be large or small, weakly acidic or basic, hydrophilic or hydrophobic, positively or negatively charged, or neutral. The unique combination of amino acid residues, their positions, sequences, structures, and properties, creates a very specific chemical environment within the active site. This specific environment is suited to bind, albeit briefly, to a specific chemical substrate (or substrates). Due to this jigsaw puzzle-like match between an enzyme and its substrates (which adapts to find the best fit between the transition state and the active site), enzymes are known for their specificity. The “best fit” results from the shape and the amino acid functional group’s attraction to the substrate. There is a specifically matched enzyme for each substrate and, thus, for each chemical reaction; however, there is flexibility as well.
The fact that active sites are so perfectly suited to provide specific environmental conditions also means that they are subject to influences by the local environment. It is true that increasing the environmental temperature generally increases reaction rates, enzyme-catalyzed or otherwise. However, increasing or decreasing the temperature outside of an optimal range can affect chemical bonds within the active site in such a way that they are less well suited to bind substrates. High temperatures will eventually cause enzymes, like other biological molecules, to denature, a process that changes the natural properties of a substance. Likewise, the pH of the local environment can also affect enzyme function. Active site amino acid residues have their own acidic or basic properties that are optimal for catalysis. These residues are sensitive to changes in pH that can impair the way substrate molecules bind. Enzymes are suited to function best within a certain pH range, and, as with temperature, extreme pH values (acidic or basic) of the environment can cause enzymes to denature.
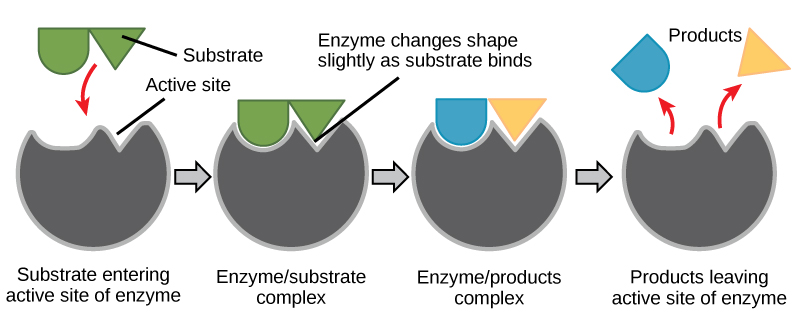
Induced Fit and Enzyme Function
For many years, scientists thought that enzyme-substrate binding took place in a simple “lock-and-key” fashion. This model asserted that the enzyme and substrate fit together perfectly in one instantaneous step. However, current research supports a more refined view called induced fit (Figure \(\PageIndex{2}\)). The induced-fit model expands upon the lock-and-key model by describing a more dynamic interaction between enzyme and substrate. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the enzyme and the transition state of the substrate. This ideal binding maximizes the enzyme’s ability to catalyze its reaction.
When an enzyme binds its substrate, an enzyme-substrate complex is formed. This complex lowers the activation energy of the reaction and promotes its rapid progression in one of many ways. On a basic level, enzymes promote chemical reactions that involve more than one substrate by bringing the substrates together in an optimal orientation. The appropriate region (atoms and bonds) of one molecule is juxtaposed to the appropriate region of the other molecule with which it must react. Another way in which enzymes promote the reaction of their substrates is by creating an optimal environment within the active site for the reaction to occur. Certain chemical reactions might proceed best in a slightly acidic or non-polar environment. The chemical properties that emerge from the particular arrangement of amino acid residues within an active site create the perfect environment for an enzyme’s specific substrates to react.
You’ve learned that the activation energy required for many reactions includes the energy involved in manipulating or slightly contorting chemical bonds so that they can easily break and allow others to reform. Enzymatic action can aid this process. The enzyme-substrate complex can lower the activation energy by contorting substrate molecules in such a way as to facilitate bond-breaking, helping to reach the transition state. Finally, enzymes can also lower activation energies by taking part in the chemical reaction itself. The amino acid residues can provide certain ions or chemical groups that actually form covalent bonds with substrate molecules as a necessary step of the reaction process. In these cases, it is important to remember that the enzyme will always return to its original state at the completion of the reaction. One of the hallmark properties of enzymes is that they remain ultimately unchanged by the reactions they catalyze. After an enzyme is done catalyzing a reaction, it releases its product(s).
Control of Metabolism Through Enzyme Regulation
It would seem ideal to have a scenario in which all of the enzymes encoded in an organism’s genome existed in abundant supply and functioned optimally under all cellular conditions, in all cells, at all times. In reality, this is far from the case. A variety of mechanisms ensure that this does not happen. Cellular needs and conditions vary from cell to cell, and change within individual cells over time. The required enzymes and energetic demands of stomach cells are different from those of fat storage cells, skin cells, blood cells, and nerve cells. Furthermore, a digestive cell works much harder to process and break down nutrients during the time that closely follows a meal compared with many hours after a meal. As these cellular demands and conditions vary, so do the amounts and functionality of different enzymes.
Since the rates of biochemical reactions are controlled by activation energy, and enzymes lower and determine activation energies for chemical reactions, the relative amounts and functioning of the variety of enzymes within a cell ultimately determine which reactions will proceed and at which rates. This determination is tightly controlled. In certain cellular environments, enzyme activity is partly controlled by environmental factors, like pH and temperature. There are other mechanisms through which cells control the activity of enzymes and determine the rates at which various biochemical reactions will occur.
Regulation of Enzymes by Molecules
Enzymes can be regulated in ways that either promote or reduce their activity. There are many different kinds of molecules that inhibit or promote enzyme function, and various mechanisms exist for doing so. In some cases of enzyme inhibition, for example, an inhibitor molecule is similar enough to a substrate that it can bind to the active site and simply block the substrate from binding. When this happens, the enzyme is inhibited through competitive inhibition, because an inhibitor molecule competes with the substrate for active site binding (Figure \(\PageIndex{3}\)). On the other hand, in noncompetitive inhibition, an inhibitor molecule binds to the enzyme in a location other than an allosteric site and still manages to block substrate binding to the active site.
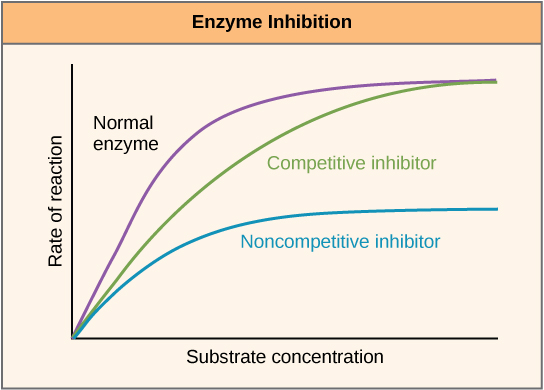
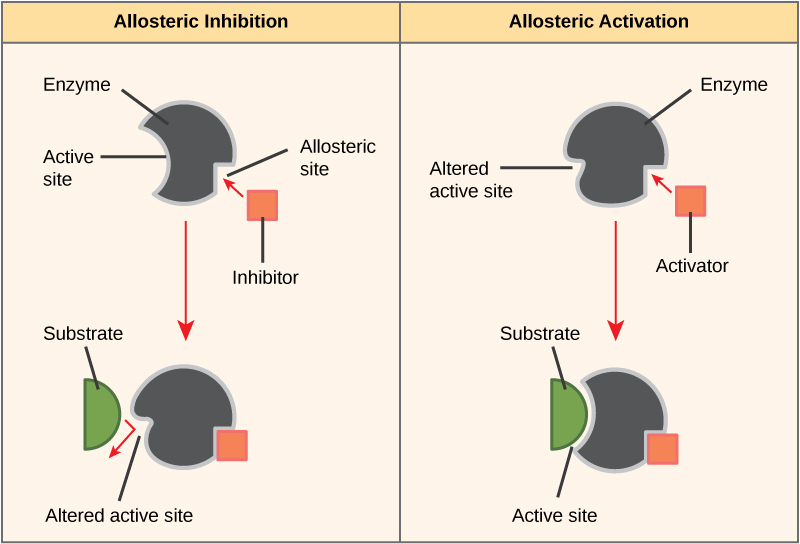
Some inhibitor molecules bind to enzymes in a location where their binding induces a conformational change that reduces the affinity of the enzyme for its substrate. This type of inhibition is called allosteric inhibition (Figure \(\PageIndex{4}\)). Most allosterically regulated enzymes are made up of more than one polypeptide, meaning that they have more than one protein subunit. When an allosteric inhibitor binds to an enzyme, all active sites on the protein subunits are changed slightly such that they bind their substrates with less efficiency. There are allosteric activators as well as inhibitors. Allosteric activators bind to locations on an enzyme away from the active site, inducing a conformational change that increases the affinity of the enzyme’s active site(s) for its substrate(s).
Everyday Connection: Drug Discovery by Looking for Inhibitors of Key Enzymes in Specific Pathways

Enzymes are key components of metabolic pathways. Understanding how enzymes work and how they can be regulated is a key principle behind the development of many of the pharmaceutical drugs (Figure \(\PageIndex{5}\)) on the market today. Biologists working in this field collaborate with other scientists, usually chemists, to design drugs.
Consider statins for example—which is the name given to the class of drugs that reduces cholesterol levels. These compounds are essentially inhibitors of the enzyme HMG-CoA reductase. HMG-CoA reductase is the enzyme that synthesizes cholesterol from lipids in the body. By inhibiting this enzyme, the levels of cholesterol synthesized in the body can be reduced. Similarly, acetaminophen, popularly marketed under the brand name Tylenol, is an inhibitor of the enzyme cyclooxygenase. While it is effective in providing relief from fever and inflammation (pain), its mechanism of action is still not completely understood.
How are drugs developed? One of the first challenges in drug development is identifying the specific molecule that the drug is intended to target. In the case of statins, HMG-CoA reductase is the drug target. Drug targets are identified through painstaking research in the laboratory. Identifying the target alone is not sufficient; scientists also need to know how the target acts inside the cell and which reactions go awry in the case of disease. Once the target and the pathway are identified, then the actual process of drug design begins. During this stage, chemists and biologists work together to design and synthesize molecules that can either block or activate a particular reaction. However, this is only the beginning: both if and when a drug prototype is successful in performing its function, then it must undergo many tests from in vitro experiments to clinical trials before it can get FDA approval to be on the market.
Many enzymes don’t work optimally, or even at all, unless bound to other specific non-protein helper molecules, either temporarily through ionic or hydrogen bonds or permanently through stronger covalent bonds. Two types of helper molecules are cofactors and coenzymes. Binding to these molecules promotes optimal conformation and function for their respective enzymes. Cofactors are inorganic ions such as iron (Fe++) and magnesium (Mg++). One example of an enzyme that requires a metal ion as a cofactor is the enzyme that builds DNA molecules, DNA polymerase, which requires bound zinc ion (Zn++) to function. Coenzymes are organic helper molecules, with a basic atomic structure made up of carbon and hydrogen, which are required for enzyme action. The most common sources of coenzymes are dietary vitamins (Figure \(\PageIndex{6}\)). Some vitamins are precursors to coenzymes and others act directly as coenzymes. Vitamin C is a coenzyme for multiple enzymes that take part in building the important connective tissue component, collagen. An important step in the breakdown of glucose to yield energy is catalysis by a multi-enzyme complex called pyruvate dehydrogenase. Pyruvate dehydrogenase is a complex of several enzymes that actually requires one cofactor (a magnesium ion) and five different organic coenzymes to catalyze its specific chemical reaction. Therefore, enzyme function is, in part, regulated by an abundance of various cofactors and coenzymes, which are supplied primarily by the diets of most organisms.
Enzyme Compartmentalization
In eukaryotic cells, molecules such as enzymes are usually compartmentalized into different organelles. This allows for yet another level of regulation of enzyme activity. Enzymes required only for certain cellular processes can be housed separately along with their substrates, allowing for more efficient chemical reactions. Examples of this sort of enzyme regulation based on location and proximity include the enzymes involved in the latter stages of cellular respiration, which take place exclusively in the mitochondria, and the enzymes involved in the digestion of cellular debris and foreign materials, located within lysosomes.
Feedback Inhibition in Metabolic Pathways
Molecules can regulate enzyme function in many ways. A major question remains, however: What are these molecules and where do they come from? Some are cofactors and coenzymes, ions, and organic molecules, as you’ve learned. What other molecules in the cell provide enzymatic regulation, such as allosteric modulation, and competitive and noncompetitive inhibition? The answer is that a wide variety of molecules can perform these roles. Some of these molecules include pharmaceutical and non-pharmaceutical drugs, toxins, and poisons from the environment. Perhaps the most relevant sources of enzyme regulatory molecules, with respect to cellular metabolism, are the products of the cellular metabolic reactions themselves. In a most efficient and elegant way, cells have evolved to use the products of their own reactions for feedback inhibition of enzyme activity. Feedback inhibition involves the use of a reaction product to regulate its own further production (Figure \(\PageIndex{7}\)). The cell responds to the abundance of specific products by slowing down production during anabolic or catabolic reactions. Such reaction products may inhibit the enzymes that catalyzed their production through the mechanisms described above.
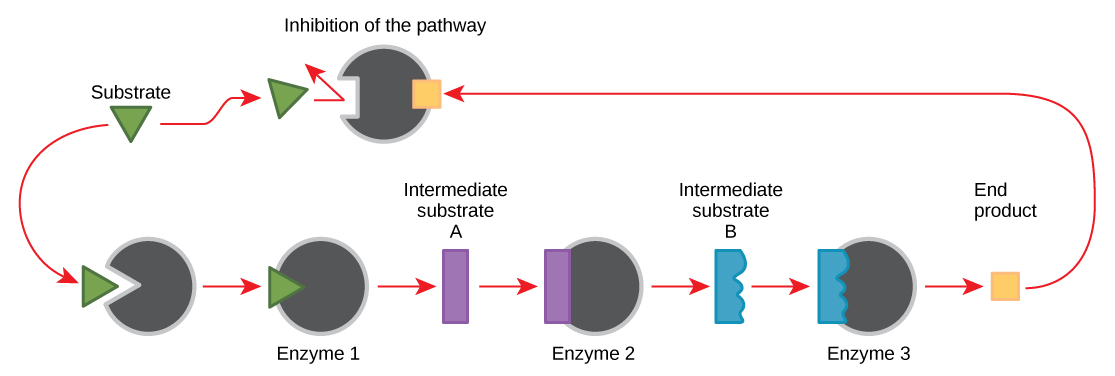
The production of both amino acids and nucleotides is controlled through feedback inhibition. Additionally, ATP is an allosteric regulator of some of the enzymes involved in the catabolic breakdown of sugar, the process that produces ATP. In this way, when ATP is abundant, the cell can prevent its further production. Remember that ATP is an unstable molecule that can spontaneously dissociate into ADP. If too much ATP were present in a cell, much of it would go to waste. On the other hand, ADP serves as a positive allosteric regulator (an allosteric activator) for some of the same enzymes that are inhibited by ATP. Thus, when relative levels of ADP are high compared to ATP, the cell is triggered to produce more ATP through the catabolism of sugar.
Summary
Enzymes are chemical catalysts that accelerate chemical reactions at physiological temperatures by lowering their activation energy. Enzymes are usually proteins consisting of one or more polypeptide chains. Enzymes have an active site that provides a unique chemical environment, made up of certain amino acid R groups (residues). This unique environment is perfectly suited to convert particular chemical reactants for that enzyme, called substrates, into unstable intermediates called transition states. Enzymes and substrates are thought to bind with an induced fit, which means that enzymes undergo slight conformational adjustments upon substrate contact, leading to full, optimal binding. Enzymes bind to substrates and catalyze reactions in four different ways: bringing substrates together in an optimal orientation, compromising the bond structures of substrates so that bonds can be more easily broken, providing optimal environmental conditions for a reaction to occur, or participating directly in their chemical reaction by forming transient covalent bonds with the substrates.
Enzyme action must be regulated so that in a given cell at a given time, the desired reactions are being catalyzed and the undesired reactions are not. Enzymes are regulated by cellular conditions, such as temperature and pH. They are also regulated through their location within a cell, sometimes being compartmentalized so that they can only catalyze reactions under certain circumstances. Inhibition and activation of enzymes via other molecules are other important ways that enzymes are regulated. Inhibitors can act competitively, noncompetitively, or allosterically; noncompetitive inhibitors are usually allosteric. Activators can also enhance the function of enzymes allosterically. The most common method by which cells regulate the enzymes in metabolic pathways is through feedback inhibition. During feedback inhibition, the products of a metabolic pathway serve as inhibitors (usually allosteric) of one or more of the enzymes (usually the first committed enzyme of the pathway) involved in the pathway that produces them.
Glossary
- active site
- specific region of the enzyme to which the substrate binds
- allosteric inhibition
- inhibition by a binding event at a site different from the active site, which induces a conformational change and reduces the affinity of the enzyme for its substrate
- coenzyme
- small organic molecule, such as a vitamin or its derivative, which is required to enhance the activity of an enzyme
- cofactor
- inorganic ion, such as iron and magnesium ions, required for optimal regulation of enzyme activity
- competitive inhibition
- type of inhibition in which the inhibitor competes with the substrate molecule by binding to the active site of the enzyme
- denature
- process that changes the natural properties of a substance
- feedback inhibition
- effect of a product of a reaction sequence to decrease its further production by inhibiting the activity of the first enzyme in the pathway that produces it
- induced fit
- dynamic fit between the enzyme and its substrate, in which both components modify their structures to allow for ideal binding
- substrate
- molecule on which the enzyme acts


