S2018_Lecture02_Reading
- Page ID
- 11382
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)General Approach to Biomolecule Types in BIS2A
Before you start
If necessary please review the Design Challenge module to review the Design Challenge rubric.
Some context and motivation
In BIS2A, we are concerned primarily with developing a functional understanding of a biological cell. In the context of a design problem, we might say that we want to solve the problem of building a cell. If we break this big task down into smaller problems, or alternatively, ask what types of things do we need to understand in order to do this, it would be reasonable to conclude that understanding what the cell is made of would be important. That said, it isn't sufficient to appreciate WHAT the cell is made of. We also need to understand the PROPERTIES of the materials that make up the cell. This requires us to dig into a little bit of chemistry—the science of the "stuff" (matter) that makes up the world we know.
This prospect of talking about molecular chemistry and thermodynamics makes some students of biology apprehensive. Hopefully, however, we will show that many of the vast number of biological processes that we care about arise directly from the chemical properties of the "stuff" that makes up life and that developing a functional understanding of some basic chemical concepts can be tremendously useful in thinking about how to solve problems in medicine, energy, and environment by attacking them at their core.
Importance of chemical composition
As a student in BIS2A, you will be asked to classify macromolecules into groups by looking at their chemical composition and, based on this composition, also infer some of the properties they might have. For example, carbohydrates typically have multiple hydroxyl groups. Hydroxyl groups are polar functional groups capable of forming hydrogen bonds. Therefore, some of the biologically relevant properties of various carbohydrates can be understood at some level by a balance between how they may tend to form hydrogen bonds with water, themselves or other molecules.
Linking structure to function
Each macromolecule plays a specific role in the overall functioning of a cell. The chemical properties and structure of a macromolecule will be directly related to its function. For example, the structure of a phospholipid can be broken down into two groups, a hydrophilic head group and a hydrophobic tail group. Each of these groups plays a role in not only the assembly of the cell membrane but also in the selectivity of substances that can/cannot cross the membrane.
The Structure of an atom
An atom is the smallest unit of matter that retains all of the chemical properties of an element. Elements are forms of matter with specific chemical and physical properties that cannot be broken down into smaller substances by ordinary chemical reactions.
The chemistry discussed in BIS2A requires us to use a model for an atom. While there are more sophisticated models, the atomic model used in this course makes the simplifying assumption that the standard atom is composed of three subatomic particles, the proton, the neutron, and the electron. Protons and neutrons have a mass of approximately one atomic mass unit (a.m.u.). One atomic mass unit is approximately 1.660538921 x 10-27kg—roughly 1/12 of the mass of a carbon atom (see table below for more precise value). The mass of an electron is 0.000548597 a.m.u. or 9.1 x 10-31kg. Neutrons and protons reside at the center of the atom in a region call the nucleus while the electrons orbit around the nucleus in zones called orbitals, as illustrated below. The only exception to this description is the hydrogen (H) atom, which is composed of one proton and one electron with no neutrons. An atom is assigned an atomic number based on the number of protons in the nucleus. Neutral carbon (C), for instance has six neutrons, six protons, and six electrons. It has an atomic number of six and a mass of slightly more than 12 a.m.u.
Table 1. Charge, mass, and location of subatomic particles
| Protons, neutrons, and electrons | ||||
| Charge | Mass (a.m.u.) | Mass (kg) | Location | |
| Proton | +1 | ~1 | 1.6726 x 10-27 | nucleus |
| Neutron | 0 | ~1 | 1.6749 x 10-27 | nucleus |
| Electron | –1 | ~0 | 9.1094 x 10-31 | orbitals |
Table 1 reports the charge and location of three subatomic particles—the neutron, proton, and electron. Atomic mass unit = a.m.u. (a.k.a. dalton [Da])—this is defined as approximately one twelfth of the mass of a neutral carbon atom or 1.660538921 x 10−27 kg. This is roughly the mass of a proton or neutron.

Figure 2. Elements, such as helium depicted here, are made up of atoms. Atoms are made up of protons and neutrons located within the nucleus and electrons surrounding the nucleus in regions called orbitals. (Note: This figure depicts a Bohr model for an atom—we could use a new open source figure that depicts a more modern model for orbitals. If anyone finds one please forward it.)
Source:(https://commons.wikimedia.org/wiki/F...um_atom_QM.svg)
By User: Yzmo (Own work) [GFDL (http://www.gnu.org/copyleft/fdl.html) or CC-BY-SA-3.0 (http://creativecommons.org/licenses/by-sa/3.0/)], via Wikimedia Commons
Relative sizes and distribution of elements
The typical atom has a radius of one to two angstroms (Å). 1Å = 1 x 10-10m. The typical nucleus has a radius of 1 x 10-5Å or 10,000 smaller than the radius of the whole atom. By analogy, a typical large exercise ball has a radius of 0.85m. If this were an atom, the nucleus would have a radius about 1/2 to 1/10 of your thinnest hair. All of that extra volume is occupied by the electrons in regions called orbitals. For an ideal atom, orbitals are probabilistically defined regions in space around the nucleus in which an electron can be expected to be found.
For additional basic information on atomic structure click here.
For additional basic information on orbitals here.
Video clips
For a review of atomic structure check out this Youtube video: atomic structure.
The properties of living and nonliving materials are determined to a large degree by the composition and organization of their constituent elements. Five elements are common to all living organisms: Oxygen (O), Carbon (C), Hydrogen (H), Phosphorous (P), and Nitrogen (N). Other elements like Sulfur (S), Calcium (Ca), Chloride (Cl), Sodium (Na), Iron (Fe), Cobalt (Co), Magnesium, Potassium (K), and several other trace elements are also necessary for life, but are typically found in far less abundance than the "top five" noted above. As a consequence, life's chemistry—and by extension the chemistry of relevance in BIS2A—largely focuses on common arrangements of and reactions between the "top five" core atoms of biology.

Figure 3. A table illustrating the abundance of elements in the human body. A pie chart illustrating the relationships in abundance between the four most common elements.
Credit: Data from Wikipedia (http://en.wikipedia.org/wiki/Abundan...mical_elements); chart created by Marc T. Facciotti
The Periodic Table
The different elements are organized and displayed in the periodic table. Devised by Russian chemist Dmitri Mendeleev (1834–1907) in 1869, the table groups elements that, due to some commonalities of their atomic structure, share certain chemical properties. The atomic structure of elements is responsible for their physical properties including whether they exist as gases, solids, or liquids under specific conditions and and their chemical reactivity, a term that refers to their ability to combine and to chemically bond with each other and other elements.
In the periodic table, shown below, the elements are organized and displayed according to their atomic number and are arranged in a series of rows and columns based on shared chemical and physical properties. In addition to providing the atomic number for each element, the periodic table also displays the element’s atomic mass. Looking at carbon, for example, its symbol (C) and name appear, as well as its atomic number of six (in the upper right-hand corner indicating the number of protons in the neutral nucleus) and its atomic mass of 12.11 (sum of the mass of electrons, protons, and neutrons).
Electronegativity
Molecules are collections of atoms that are associated with one another through bonds. It is reasonable to expect—and the case empirically—that different atoms will exhibit different physical properties, including abilities to interact with other atoms. One such property, the tendency of an atom to attract electrons, is described by the chemical concept and term, electronegativity. While several methods for measuring electronegativity have been developed, the one most commonly taught to biologists is the one created by Linus Pauling.
A description of how Pauling electronegativity can be calculated is beyond the scope of BIS2A. What is important to know, however, is that electronegativity values have been experimentally and/or theoretically determined for nearly all elements in the periodic table. The values are unitless and are reported relative to the standard reference, hydrogen, whose electronegativity is 2.20. The larger the electronegativity value, the greater tendency an atom has to attract electrons. Using this scale, the electronegativity of different atoms can be quantitatively compared. For instance, by using Table 1 below, you could report that oxygen atoms (O) are more electronegative than phosphorous atoms (P).
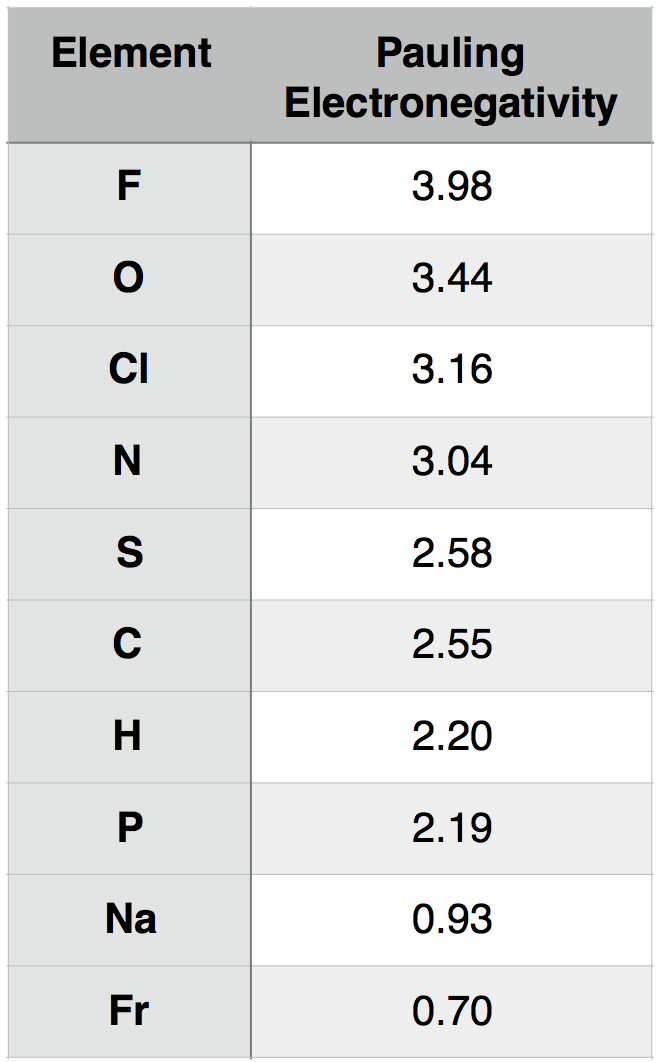
Table 1. Pauling electronegativity values for select elements of relevance to BIS2A as well as elements at the two extremes (highest and lowest) of the electronegativity scale.
Attribution: Marc T. Facciotti (original work)
The utility of the Pauling electronegativity scale in BIS2A is to provide a chemical basis for explaining the types of bonds that form between the commonly occurring elements in biological systems and to explain some of the key interactions that we observe routinely. We develop our understanding of electronegativity-based arguments about bonds and molecular interactions by comparing the electronegativities of two atoms. Recall, the larger the electronegativity, the stronger the "pull" an atom exerts on nearby electrons.
We can consider, for example, the common interaction between oxygen (O) and hydrogen (H). Let us assume that O and H are interacting (forming a bond) and write that interaction as O-H, where the dash between the letters represents the interaction between the two atoms. To understand this interaction better, we can compare the relative electronegativity of each atom. Examining the table above, we see that O has an electronegativity of 3.44, and H has an electronegativity of 2.20.
Based on the concept of electronegativity as we now understand it, we can surmise that the oxygen (O) atom will tend to "pull" the electrons away from the hydrogen (H) when they are interacting. This will give rise to a slight but significant negative charge around the O atom (due to the higher tendency of the electrons to be associated with the O atom). This also results in a slight positive charge around the H atom (due to the decrease in the probability of finding an electron nearby). Since the electrons are not distributed evenly between the two atoms AND, by consequence, the electric charge is also not evenly distributed, we describe this interaction or bond as polar. There are two poles in effect: the negative pole near the oxygen and the positive pole near the hydrogen.
To extend the utility of this concept, we can now ask how an interaction between oxygen (O) and hydrogen (H) differs from an interaction between sulfur (S) and hydrogen (H). That is, how does O-H differ from S-H? If we examine the table above, we see that the difference in electronegativity between O and H is 1.24 (3.44 - 2.20 = 1.24) and that the difference in electronegativity between S and H is 0.38 (2.58 – 2.20 = 0.38). We can therefore conclude that an O-H bond is more polar than an S-H bond. We will discuss the consequences of these differences in subsequent chapters.

Figure 2. The periodic table with the electronegativities of each atom listed.
Attribution: By DMacks (https://en.wikipedia.org/wiki/Electronegativity) [CC BY-SA 3.0 (http://creativecommons.org/licenses/by-sa/3.0)], via Wikimedia Commons
An examination of the periodic table of the elements (Figure 2) illustrates that electronegativity is related to some of the physical properties used to organize the elements into the table. Certain trends are apparent. For instance, those atoms with the largest electronegativity tend to reside in the upper right hand corner of the periodic table, such as Fluorine (F), Oxygen (O) and Chlorine (Cl), while elements with the smallest electronegativity tend to be found at the other end of the table, in the lower left, such as Francium (Fr), Cesium (Cs) and Radium (Ra).
More information on electronegativity can be found in the LibreTexts.
The main use of the concept of electronegativity in BIS2A will therefore be to provide a conceptual grounding for discussing the different types of chemical bonds that occur between atoms in nature. We will focus primarily on three types of bonds: Ionic Bonds, Covalent Bonds and Hydrogen Bonds.
Bond types
In BIS2A, we focus primarily on three different bond types: ionic bonds, covalent bonds, and hydrogen bonds. We expect students to be able to recognize each different bond type in molecular models. In addition, for commonly seen bonds in biology, we expect student to provide a chemical explanation, rooted in ideas like electronegativity, for how these bonds contribute to the chemistry of biological molecules.
Ionic bonds
Ionic bonds are electrostatic interactions formed between ions of opposite charges. For instance, most of us know that in sodium chloride (NaCl) positively charged sodium ions and negatively charged chloride ions associate via electrostatic (+ attracts -) interactions to make crystals of sodium chloride, or table salt, creating a crystalline molecule with zero net charge. The origins of these interactions may arise from the association of neutral atoms whose difference in electronegativities is sufficiently high. Take the example above. If we imagine that a neutral sodium atom and a neutral chlorine atom approach one another, it is possible that at close distances, due to the relatively large difference in electronegativity between the two atoms, that an electron from the neutral sodium atom is transferred to the neutral chlorine atom, resulting in a negatively charged chloride ion and a positively charged sodium ion. These ions can now interact via an ionic bond.

Figure 1. The formation of an ionic bond between sodium and chlorine is depicted. In panel A, a sufficient difference in electronegativity between sodium and chlorine induces the transfer of an electron from the sodium to the chlorine, forming two ions, as illustrated in panel B. In panel C, the two ions associate via an electrostatic interaction. Attribution: By BruceBlaus (own work) [CC BY-SA 4.0 (http://creativecommons.org/licenses/by-sa/4.0)], via Wikimedia Commons
This movement of electrons from one atom to another is referred to as electron transfer. In the example above, when sodium loses an electron, it now has 11 protons, 11 neutrons, and 10 electrons, leaving it with an overall charge of +1 (summing charges: 11 protons at +1 charge each and 10 electrons at -1 charge each = +1). Once charged, the sodium atom is referred to as a sodium ion. Likewise, based on its electronegativity, a neutral chlorine (Cl) atom tends to gain an electron to create an ion with 17 protons, 17 neutrons, and 18 electrons, giving it a net negative (–1) charge. It is now referred to as a chloride ion.
We can interpret the electron transfer above using the concept of electronegativity. Begin by comparing the electronegativities of sodium and chlorine by examining the periodic table of elements below. We see that chlorine is located in the upper-right corner of the table, while sodium is in the upper left. Comparing the electronegativity values of chlorine and sodium directly, we see that the chlorine atom is more electronegative than is sodium. The difference in the electronegativity of chlorine (3.16) and sodium (0.93) is 2.23 (using the scale in the table below). Given that we know an electron transfer will take place between these two elements, we can conclude that differences in electronegativities of ~2.2 are large enough to cause an electron to transfer between two atoms and that interactions between such elements are likely through ionic bonds.

Figure 2. The periodic table of the elements listing electronegativity values for each element. The elements sodium and chlorine are boxed with a teal boundary. Attribution: By DMacks (https://en.wikipedia.org/wiki/Electronegativity) [CC BY-SA 3.0 (http://creativecommons.org/licenses/by-sa/3.0)], via Wikimedia Commons—Modified by Marc T. Facciotti
Note: possible discussion
The atoms in a 5 in. x 5 in. brick of table salt (NaCl) sitting on your kitchen counter are held together almost entirely by ionic bonds. Based on that observation, how would you characterize the strength of ionic bonds?
Now consider that same brick of table salt after having been thrown into an average backyard swimming pool. After a couple of hours, the brick would be completely dissolved, and the sodium and chloride ions would be uniformly distributed throughout the pool. What might you conclude about the strength of ionic bonds from this observation?
Propose a reason why NaCl's ionic bonds in air might be behaving differently than those in water? What is the significance of this to biology?
For additional information:
Check out the link from the Khan Academy on ionic bonds.
Covalent bonds
We can also invoke the concept of electronegativity to help describe the interactions between atoms that have differences in electronegativity too small for the atoms to form an ionic bond. These types of interactions often result in a bond called a covalent bond. In these bonds, electrons are shared between two atoms—in contrast to an ionic interaction in which electrons remain on each atom of an ion or are transferred between species that have highly different electronegativities.
We start exploring the covalent bond by looking at an example where the difference in electronegativity is zero. Consider a very common interaction in biology, the interaction between two carbon atoms. In this case, each atom has the same electronegativity, 2.55; the difference in electronegativity is therefore zero. If we build our mental model of this interaction using the concept of electronegativity, we realize that each carbon atom in the carbon-carbon pair has the same tendency to "pull" electrons to it. In this case, when a bond is formed, neither of the two carbon atoms will tend to "pull" (a good anthropomorphism) electrons from the other. They will "share" (another anthropomorphism) the electrons equally, instead.
Aside: bounding example
The two examples above—(1) the interaction of sodium and chlorine, and (2) the interaction between two carbon atoms—frame a discussion by "bounding," or asymptotic analysis (see earlier reading). We examined what happens to a physical system when considering two extremes. In this case, the extremes were in electronegativity differences between interacting atoms. The interaction of sodium and chlorine illustrated what happens when two atoms have a large difference in electronegativities, and the carbon-carbon example illustrated what happens when that difference is zero. Once we create those mental goal posts describing what happens at the extremes, it is then easier to imagine what might happen in between—in this case, what happens when the difference in electronegativity is between 0 and 2.2. We do that next.
When the sharing of electrons between two covalently bonded atoms is nearly equal, we call these bonds nonpolar covalent bonds. If by contrast, the sharing of electrons is not equal between the two atoms (likely due to a difference in electronegativities between the atoms), we call these bonds polar covalent bonds.
In a polar covalent bond, the electrons are unequally shared by the atoms and are attracted to one nucleus more than to the other. Because of the unequal distribution of electrons between atoms in a polar covalent bond, a slightly positive (indicated by δ+) or slightly negative (indicated by δ–) charge develops at each pole of the bond. The slightly positive (δ+) charge will develop on the less electronegative atom, as electrons get pulled more towards the slightly more electronegative atom. A slightly negative (δ–) charge will develop on the more electronegative atom. Since there are two poles (the positive and negative poles), the bond is said to possess a dipole.
Examples of nonpolar covalent and polar covalent bonds in biologically relevant molecules
Nonpolar covalent bonds
Molecular oxygen
Molecular oxygen (O2) is made from an association between two atoms of oxygen. Since the two atoms share the same electronegativity, the bonds in molecular oxygen are nonpolar covalent.
Methane
Another example of a nonpolar covalent bond is the C-H bond found in the methane gas (CH4). Unlike the case of molecular oxygen where the two bonded atoms share the same electronegativity, carbon and hydrogen do not have the same electronegativity; C = 2.55 and H = 2.20—the difference in electronegativity is 0.35.

Figure 3. Molecular line drawings of molecular oxygen, methane, and carbon dioxide. Attribution: Marc T. Facciotti (own work)
Some of you may now be confused. If there is a difference in electronegativity between the two atoms, is the bond not by definition polar? The answer is both yes and no and depends on the definition of polar that the speaker/writer is using. Since this is an example of how taking shortcuts in the use of specific vocabulary can sometimes lead to confusion, we take a moment to discuss this here. See the mock exchange between a student and an instructor below for clarification:
1. Instructor: "In biology, we often say that the C-H bond is nonpolar."
2. Student: "But there is an electronegativity difference between C and H, so it would appear that C should have a slightly stronger tendency to attract electrons. This electronegativity difference should create a small, negative charge around the carbon and a small, positive charge around the hydrogen."
3. Student: "Since there is a differential distribution of charge across the bond, it would seem that, by definition, this should be considered a polar bond."
4. Instructor: "In fact, the bond does have some small polar character."
5. Student: "So, then it's polar? I'm confused."
6. Instructor: "It has some small amount of polar character, but it turns out that for most of the common chemistry that we will encounter that this small amount of polar character is insufficient to lead to "interesting" chemistry. So, while the bond is, strictly speaking, slightly polar, from a practical standpoint it is effectively nonpolar. We therefore call it nonpolar."
7. Student: "That's needlessly confusing; how am I supposed to know when you mean strictly 100% nonpolar, slightly polar, or functionally polar when you use the same word to describe two of those three things?"
8. Instructor: "Yup, it sucks. The fix is that I need to be as clear as I can when I talk with you about how I am using the term "polarity." I also need to inform you that you will find this shortcut (and others) used when you go out into the field, and I encourage you to start learning to recognize what is intended by the context of the conversation.
A real-world analogy of this same problem might be the use of the word "newspaper". It can be used in a sentence to refer to the company that publishes some news, OR it can refer to the actual item that the company produces. In this case, the disambiguation is easily made by native English speakers, as they can determine the correct meaning from the context; non-native speakers may be more confused. Don't worry; as you see more examples of technical word use in science, you'll learn to read correct meanings from contexts too."
Aside:
How large should the difference in electronegativity be in order to create a bond that is "polar enough" that we decide to call it polar in biology? Of course, the exact value depends on a number of factors, but as a loose rule of thumb, we sometimes use a difference of 0.4 as a guesstimate.
This extra information is purely for your information. You will not be asked to assign polarity based on this criteria in BIS2A. You should, however, appreciate the concept of how polarity can be determined by using the concept of electronegativity. You should also appreciate the functional consequences of polarity (more on this in other sections) and the nuances associated with these terms (such as those in the discussion above).
Polar covalent bonds
The polar covalent bond can be illustrated by examining the association between O and H in water (H2O). Oxygen has an electronegativity of 3.44, while hydrogen has an electronegativity of 2.20. The difference in electronegativity is 1.24. It turns out that this size of electronegativity difference is large enough that the dipole across the molecule contributes to chemical phenomenon of interest.
This is a good point to mention another common source of student confusion regarding the use of the term polar. Water has polar bonds. This statement refers specifically to the individual O-H bonds. Each of these bonds has a dipole. However, students will also hear that water is a polar molecule. This is also true. This latter statement is referring to the fact that the sum of the two bond dipoles creates a dipole across the whole molecule. A molecule may be nonpolar but still have some polar bonds.

Figure 4. A water molecule has two polar O-H bonds. Since the distribution of charge in the molecule is asymmetric (due to the number and relative orientations of the bond dipoles), the molecule is also polar. The element name and electronegativities are reported in the respective sphere. Attribution: Marc T. Facciotti (own work)
For additional information, view this short video to see an animation of ionic and covalent bonding.
The continuum of bonds between covalent and ionic
The discussion of bond types above highlights that in nature you will see bonds on a continuum from completely nonpolar covalent to purely ionic, depending on the atoms that are interacting. As you proceed through your studies, you will further discover that in larger, multi-atom molecules, the localization of electrons around an atom is also influenced by multiple factors. For instance, other atoms that are also bonded nearby will exert an influence on the electron distribution around a nucleus in a way that is not easily accounted for by invoking simple arguments of pairwise comparisons of electronegativity. Local electrostatic fields produced by other non-bonded atoms may also have an influence. Reality is always more complicated than are our models. However, if the models allow us to reason and predict with "good enough" precision or to understand some key underlying concepts that can be extended later, they are quite useful.
Key bonds in BIS2A
In BIS2A, we are concerned with the chemical behavior of and bonds between atoms in biomolecules. Fortunately, biological systems are composed of a relatively small number of common elements (e.g., C, H, N, O, P, S, etc.) and some key ions (e.g., Na+, Cl-, Ca2+, K+, etc.). Start recognizing commonly occurring bonds and the chemical properties that we often see them showing. Some common bonds include C-C, C-O, C-H, N-H, C=O, C-N, P-O, O-H, S-H, and some variants. These will be discussed further in the context of functional groups. The task is not as daunting as it seems.
Note Common Point of student confusion
In this reading we have been talking about the polarity of bonds. That is, we have been learning how to describe the polarity of a single bond joining two atoms (i.e. how are the electrons shared between two atoms distributed about the respective nuclei?). In biology we also sometimes talk about the polarity of a molecule. The polarity of a molecule is different than the polarity of a bond within the molecule. The latter is asking whether the whole molecule has a net dipole. The molecule's dipole can be roughly thought of as the sum of all of its bond dipoles. For example, let us examine a molecule of CO2 depicted in the figure above. If we ask whether one of the C=O bonds is polar we would conclude that it is since the oxygen is significantly more electronegative that the carbon to which it is covalently bonded. However, if we ask whether the molecule O=C=O is polar we would concluded that it is not. Why? Look at the figure of CO2 above. Each CO bond has a dipole. However, these two dipoles are pointed in directly opposite directions. If we add these two bond dipoles together to get the net dipole of the molecule we get nothing - the two bond dipoles "cancel" one another out. By contrast, if we examine the structure of water above, we also see that each O-H bond has a dipole. In this case when we ask whether the molecule has a net dipole (done by adding the bond dipoles together) we see that the answer is yes. The sum of the the two bond dipoles still yields a net dipole moment. We therefore say that this molecule is polar. We can do this same exercise for parts of molecules so long as we define what specific part we are looking at.



