2017_SS1_Lecture_12
( \newcommand{\kernel}{\mathrm{null}\,}\)
Protein Synthesis
Introduction
The process of translation in biology is the decoding an mRNA message into a polypeptide product. Put another way, a message written in the chemical language of nucleotides is "translated" into the chemical language of amino acids. Amino acids are linearly strung together via covalent bonds (called peptide bonds) between amino and carboxyl termini of adjacent amino acids. The decoding and "linking" process is catalyzed by a ribonucleoprotein complex called the ribosomes and can result in chains of amino acids of lengths ranging from tens to more than 1,000.
The resulting proteins are so important to the cell that their synthesis consumes more of a cell’s energy than any other metabolic process. Like DNA replication and transcription, translation is a complex molecular process that we can approach using both the Energy Story and Design Challenge rubrics. Describing the overall process, or steps in the process, requires the accounting of the matter and energy before the process and after the process and a description of how that matter is transformed and energy transferred during the process. From a Design Challenge standpoint, we can - even before digging any further into what is or is not understood about translation - try to infer some of the basic questions that we will need to answer regarding this process.
Let us start by considering the basic problem. We have a strand of RNA (called mRNA) and a bunch of amino acids and we need to somehow design a machine that will:
(a) decode the chemical language of nucleotides into the language of amino acids,
(b) join amino acids in a very specific manner,
(c) complete this process with reasonable accuracy, and
(d) do this at a reasonable speed. Reasonable, is of course defined by natural selection.
As before, we can identify subproblems
(a) How does our molecular machine determine where and when to start working?
(b) How does the molecular machine coordinate decoding and bond formations?
(c) where does the energy for this process come from and how much?
(d) how does the machine know where to stop?
Other questions and functional problems/challenges will certainly arise as we dig deeper.
The point, as always, is that even without knowing any specifics about translation we can use our imaginations, curiosity and common sense to imagine some requirements for the process that we will need to learn more about. Understanding these questions as the context for what follows is key.
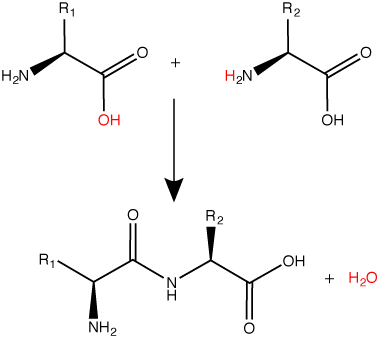
A peptide bond links the carboxyl end of one amino acid with the amino end of another, expelling one water molecule. The R1 and R2 designation refer to side chain of amino acid the two amino acids.
Attribution: Marc T. Facciotti (original work).
Protein Synthesis Machinery
The components that go into the process
Many different molecules and macromolecules contribute to the process of translation. While the exact composition of "the players" in the process may vary from species to species - for instance, ribosomes may consist of different numbers of rRNAs (ribosomal RNAs) and polypeptides depending on the organism - the general functions of the protein synthesis machinery are comparable from bacteria to human cells. We focus on these similarities. At a minimum, translation requires an mRNA template, amino acids, ribosomes, tRNAs, an energy source, and various additional accessory enzymes and small molecules.
Reminder: Amino acids
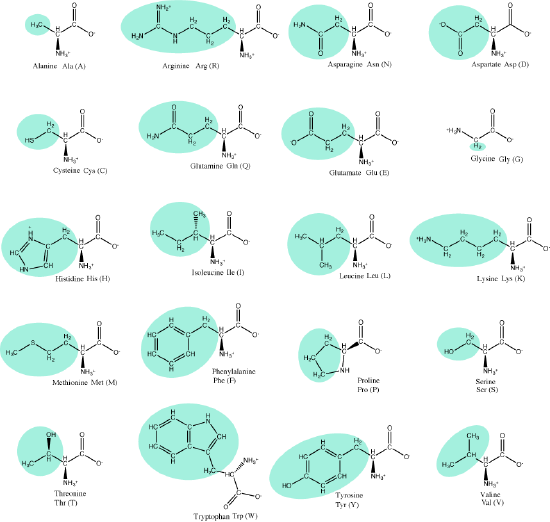
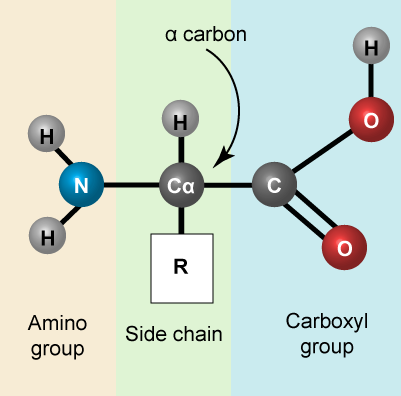
Let us simply recall that the basic structure of amino acids is composed of a backbone composed of an amino group, a central carbon (called the α-carbon), and a carboxyl group. Attached to the α-carbon is a variable group that helps determine some of the chemical properties and reactivity of the amino acid.
Ribosomes
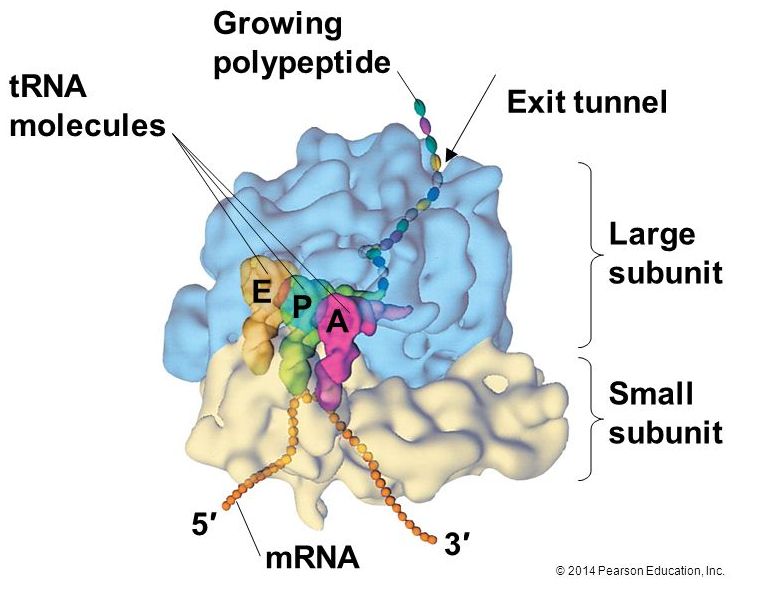
A ribosome is a complex macromolecule composed of structural and catalytic rRNAs, and many distinct polypeptides. As we start to try thinking about energy accounting in the cell it is worth noting that ribosomes do not come "free". Even before an mRNA is translated, a cell must invest energy to build each of its ribosomes. In E. coli, there are between 10,000 and 70,000 ribosomes present in each cell at any given time.
Ribosomes exist in the cytoplasm in bacteria and archaea and in the cytoplasm and on the rough endoplasmic reticulum in eukaryotes. Mitochondria and chloroplasts also have their own ribosomes in the matrix and stroma, which look more similar to bacterial ribosomes (and have similar drug sensitivities), than the ribosomes just outside their outer membranes in the cytoplasm. Ribosomes dissociate into large and small subunits when they are not synthesizing proteins and reassociate during the initiation of translation. In E. coli, the small subunit is described as 30S, and the large subunit is 50S. Mammalian ribosomes have a small 40S subunit and a large 60S subunit. The small subunit is responsible for binding the mRNA template, whereas the large subunit sequentially binds tRNAs. Each mRNA molecule is simultaneously translated by many ribosomes, all synthesizing protein in the same direction: reading the mRNA from 5' to 3' and synthesizing the polypeptide from the N terminus to the C terminus. The complete mRNA/poly-ribosome structure is called a polysome.
tRNAs
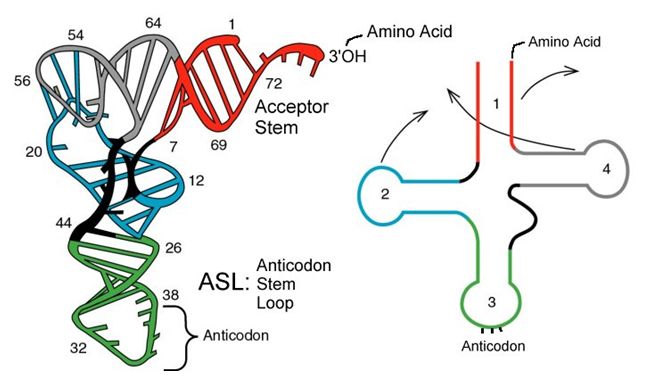
tRNAs are structural RNA molecules that were transcribed from genes. Depending on the species, 40 to 60 types of tRNAs exist in the cytoplasm. Serving as adaptors, specific tRNAs bind to sequences on the mRNA template and add the corresponding amino acid to the polypeptide chain. Therefore, tRNAs are the molecules that actually “translate” the language of RNA into the language of proteins.
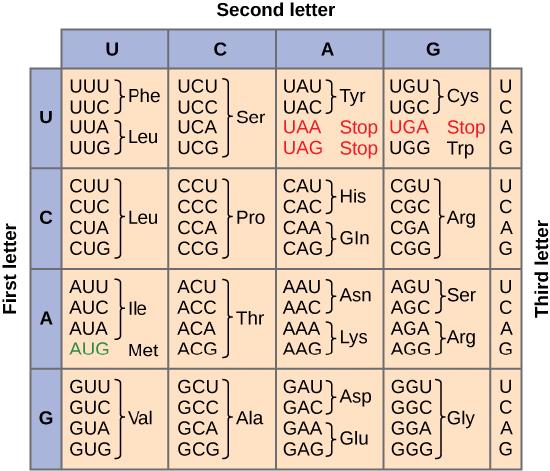
Of the 64 possible mRNA codons—or triplet combinations of A, U, G, and C, three specify the termination of protein synthesis and 61 specify the addition of amino acids to the polypeptide chain. Of these 61, one codon (AUG) also encodes the initiation of translation. Each tRNA anticodon can base pair with one of the mRNA codons and add an amino acid or terminate translation, according to the genetic code. For instance, if the sequence CUA occurred on an mRNA template in the proper reading frame, it would bind a tRNA expressing the complementary sequence, GAU, which would be linked to the amino acid leucine.
Aminoacyl tRNA Synthetases
The process of pre-tRNA synthesis by RNA polymerase III only creates the RNA portion of the adaptor molecule. The corresponding amino acid must be added later, once the tRNA is processed and exported to the cytoplasm. Through the process of tRNA “charging,” each tRNA molecule is linked to its correct amino acid by a group of enzymes called aminoacyl tRNA synthetases. At least one type of aminoacyl tRNA synthetase exists for each of the 20 amino acids; the exact number of aminoacyl tRNA synthetases varies by species. These enzymes first bind and hydrolyze ATP to catalyze a high-energy bond between an amino acid and adenosine monophosphate (AMP); a pyrophosphate molecule is expelled in this reaction. The activated amino acid is then transferred to the tRNA, and AMP is released.
The Mechanism of Protein Synthesis
Just as with mRNA synthesis, protein synthesis can be divided into three phases: initiation, elongation, and termination. The process of translation is similar in bacteria, archaea and eukaryotes.
Translation Initiation
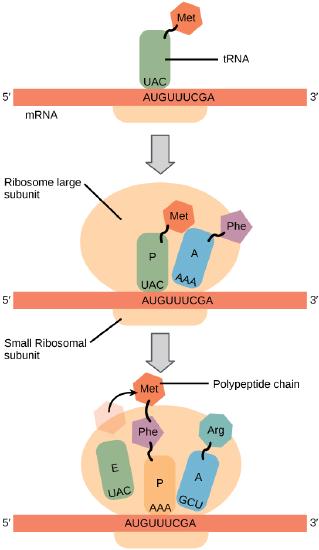
In general, protein synthesis begins with the formation of an initiation complex. The small ribosomal subunit will bind to the mRNA at the ribosomal binding site. Soon after, the methionine-tRNA will bind to the AUG start codon (through complementary binding with its anticodon). This complex is then joined by large ribosomal subunit. This initiation complex then recruits the second tRNA and thus translation begins.
Bacterial vs Eukaryotic initiation
In E. coli mRNA, a sequence upstream of the first AUG codon, called the Shine-Dalgarno sequence (AGGAGG), interacts with a rRNA molecule. This interaction anchors the 30S ribosomal subunit at the correct location on the mRNA template. Stop for a moment to appreciate the repetition of a mechanism you've encountered before. In this case, getting a protein complex to associate - in proper register - with a nucleic acid polymer is accomplished by aligning two antiparallel strands of complementary nucleotides with one another. We also saw this in the function of telomerase.
Instead of binding at the Shine-Dalgarno sequence, the eukaryotic initiation complex recognizes the 7-methylguanosine cap at the 5' end of the mRNA. A cap-binding protein (CBP) assists the movement of the ribosome to the 5' cap. Once at the cap, the initiation complex tracks along the mRNA in the 5' to 3' direction, searching for the AUG start codon. Many eukaryotic mRNAs are translated from the first AUG, but this is not always the case. According to Kozak’s rules, the nucleotides around the AUG indicate whether it is the correct start codon. Kozak’s rules state that the following consensus sequence must appear around the AUG of vertebrate genes: 5'-gccRccAUGG-3'. The R (for purine) indicates a site that can be either A or G, but cannot be C or U. Essentially, the closer the sequence is to this consensus, the higher the efficiency of translation.
Translation Elongation
During translation elongation, the mRNA template provides specificity. As the ribosome moves along the mRNA, each mRNA codon comes into 'view', and specific binding with the corresponding charged tRNA anticodon is ensured. If mRNA were not present in the elongation complex, the ribosome would bind tRNAs nonspecifically. Note again the use of base pairing between two antiparallel strands of complementary nucleotides to bring and keep our molecular machine in register and in this case also to accomplish the job of "translating" between the language of nucleotides and amino acids.
The large ribosomal subunit consists of three compartments: the A site binds incoming charged tRNAs (tRNAs with their attached specific amino acids), the P site binds charged tRNAs carrying amino acids that have formed bonds with the growing polypeptide chain but have not yet dissociated from their corresponding tRNA, and the E site which releases dissociated tRNAs so they can be recharged with another free amino acid.
Elongation proceeds with charged tRNAs entering the A site and then shifting to the P site followed by the E site with each single-codon “step” of the ribosome. Ribosomal steps are induced by conformational changes that advance the ribosome by three bases in the 3' direction. The energy for each step of the ribosome is donated by an elongation factor that hydrolyzes GTP. Peptide bonds form between the amino group of the amino acid attached to the A-site tRNA and the carboxyl group of the amino acid attached to the P-site tRNA. The formation of each peptide bond is catalyzed by peptidyl transferase, an RNA-based enzyme that is integrated into the 50S ribosomal subunit. The energy for each peptide bond formation is derived from GTP hydrolysis, which is catalyzed by a separate elongation factor. The amino acid bound to the P-site tRNA is also linked to the growing polypeptide chain. As the ribosome steps across the mRNA, the former P-site tRNA enters the E site, detaches from the amino acid, and is expelled. The ribosome moves along the mRNA, one codon at a time, catalyzing each process that occurs in the three sites. With each step, a charged tRNA enters the complex, the polypeptide becomes one amino acid longer, and an uncharged tRNA departs. Amazingly, this process occurs rapidly in the cell, the E. coli translation apparatus takes only 0.05 seconds to add each amino acid, meaning that a 200-amino acid polypeptide could be translated in just 10 seconds.
Suggested discussion
Many antibiotics inhibit bacterial protein synthesis. For example, tetracycline blocks the A site on the bacterial ribosome, and chloramphenicol blocks peptidyl transfer. What specific effect would you expect each of these antibiotics to have on protein synthesis?
The Genetic Code
To summarize what we know to this point, the cellular process of transcription generates messenger RNA (mRNA), a mobile molecular copy of one or more genes with an alphabet of A, C, G, and uracil (U). Translation of the mRNA template converts nucleotide-based genetic information into a protein product. Protein sequences consist of 20 commonly occurring amino acids; therefore, it can be said that the protein alphabet consists of 20 letters. Each amino acid is defined by a three-nucleotide sequence called the triplet codon. The relationship between a nucleotide codon and its corresponding amino acid is called the genetic code. Given the different numbers of “letters” in the mRNA and protein “alphabets,” means that there are a total of 64 (4 × 4 × 4) possible codons; therefore, a given amino acid (20 total) must be encoded for by more than one codon.
Three of the 64 codons terminate protein synthesis and release the polypeptide from the translation machinery. These triplets are called stop codons. Another codon, AUG, also has a special function. In addition to specifying the amino acid methionine, it also serves as the start codon to initiate translation. The reading frame for translation is set by the AUG start codon near the 5' end of the mRNA. The genetic code is universal. With a few exceptions, virtually all species use the same genetic code for protein synthesis, which is powerful evidence that all life on Earth shares a common origin.
Redundant, not Ambiguous
The information in the genetic code is redundant. Multiple codons code for the same amino acid. For example, using the chart above, you can find 4 different codons that code for Valine, likewise, there are two codons that code for Leucine, etc. But the code is not ambiguous, meaning, that if you were given a codon you would know definitively which amino acid it is coding for, a codon will only code for a specific amino acid. For example, GUU will always code for Valine, and AUG will always code for Methionine. This is important, you will be asked to translate an mRNA into a protein using a codon chart like the one shown above.
Translation Termination
Termination of translation occurs when a stop codon (UAA, UAG, or UGA) is encountered. When the ribosome encounters the stop codon no tRNA enters into the A site. Instead a protein know as a release factor binds to the complex. This interaction destabilizes the translation machinery, causing the release of the polypeptide and the dissociation of the ribosome subunits from the mRNA. After many ribosomes have completed translation, the mRNA is degraded so the nucleotides can be reused in another transcription reaction.
Suggested discussion
What are the benefits and drawbacks to translating a single mRNA multiple times?
Coupling between Transcription and Translation
As discussed previously, bacteria and archaea do not need to transport their RNA transcripts between a membrane bound nucleous and the cytoplasm. The RNA polymerase is therefore transcribing RNA directly into the cytoplasm. Here ribosomes can bind to the RNA and begin the process of translation, in some instances while transciption is still occurring. The coupling of these two processes, and even mRNA degradation, is facilitated not only because transcription and translation happen in the same compartment but also because both of the processes happen in the same direction - synthesis of the RNA transcript happens in the 5' to 3' direction and translation reads the transcript in the 5' to 3' direction. This "coupling" of transcription with translation occurs in both bacteria and archaea and is, in fact, essential for proper gene expression in some instances.
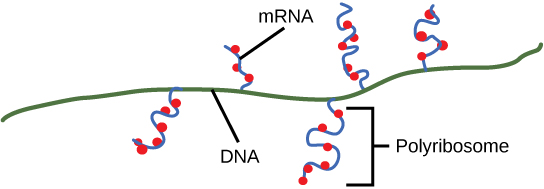
Protein Sorting
In context of a protein synthesis Design Challenge we can also raise the question/problem of how proteins get to where they are supposed to go. We know that some proteins are destined for the plasma membrane, others in eukaryotic cells need to be directed to various organelles, some proteins, like hormones or nutrient scavenging proteins, are intended to be secreted by cells while others may need to be directed to parts of the cytosol to serve structural roles. How does this happen?
Since various mechanisms have been uncovered, the details of this process are not easily summarized in a brief paragraph or two. However, some key common elements of all mechanisms can be mentioned. First, is the need for a specific "tag" that can provide some molecular information about where the protein of interest is destined. This tag usually takes the form of a short string of amino acids - a so called signal peptide - that can encode information about where the protein is intended to end up. The second required component of the protein sorting machinery must be a system to actually read and sort the proteins. In bacterial and archaeal systems this usually consists of proteins that can identify the signal peptide during translation, bind to it, and direct the synthesis of the nascent protein to the plasma membrane. In eukaryotic systems, the sorting is by necessity more complex, and involves a rather elaborate set of mechanisms of signal recognition, protein modification, and trafficking of vesicles between organelles or the membrane. These biochemical steps are initiated in the endoplasmic reticulum and further "refined" in the Golgi apparatus where proteins are modified and packaged into vesicles bound for various parts of the cell.
Some of the various specific mechanisms may be discussed by your instructor in class. The key for all students it so appreciate the problem and to have a general idea of the high-level requirements that cells have adopted to solve them.
Post-translational Protein Modification
After translation individual amino acids may be chemically modified. These modifications add chemical variation and new properties that are rooted in the chemistries of the functional groups that are being added. Common modifications include phosphate groups, methyl, acetate, and amide groups. Some proteins, typically targeted to membranes will be lipidated - a lipid will be added. Other proteins will be glycosylated - a sugar will be added. Another common post-translational modification is cleavage or linking of parts of the protein itself. Signal-peptides may be cleaved, parts may be excised from the middle of the protein, or new covalent linkages may be made between cysteine or other amino acid side chains. Nearly all modifications will be catalyzed by enzymes and all change the functional behavior of the protein.
Section Summary
mRNA is used to synthesize proteins by the process of translation. The genetic code is the correspondence between the three-nucleotide mRNA codon and an amino acid. The genetic code is “translated” by the tRNA molecules, which associate a specific codon with a specific amino acid. The genetic code is degenerate because 64 triplet codons in mRNA specify only 20 amino acids and three stop codons. This means that more than one codon corresponds to an amino acid. Almost every species on the planet uses the same genetic code.
The players in translation include the mRNA template, ribosomes, tRNAs, and various enzymatic factors. The small ribosomal subunit binds to the mRNA template. Translation begins at the initiating AUG on the mRNA. The formation of bonds occurs between sequential amino acids specified by the mRNA template according to the genetic code. The ribosome accepts charged tRNAs, and as it steps along the mRNA, it catalyzes bonding between the new amino acid and the end of the growing polypeptide. The entire mRNA is translated in three-nucleotide “steps” of the ribosome. When a stop codon is encountered, a release factor binds and dissociates the components and frees the new protein.
Introduction to gene regulation
Regulation is all about decision making. Gene regulation is, therefore, all about understanding how cells make decisions about which genes to turn on, turn off or to tune up or tune down. In the following section we discuss some of the fundamental mechanisms and principles used by cells to regulate gene expression in response to changes in cellular or external factors. This biology is important for understanding how cells adjust changing environments, including how some cells, in multicellular organisms, decide to become specialized for certain functions (e.g. tissues).
Since the subject of regulation is both a very deep and broad topic of study in biology, in Bis2a we don't try to cover every detail - there are simply way too many. Rather, as we have done for all other topics, we try to focus on (a) outlining some of the core logical constructs and questions that you must have when you approach ANY scenario involving regulation, (b) learning some common vocabulary and ubiquitous mechanisms and (c) examining a few concrete examples that illustrate the points made in a and b.
Gene Expression
Introduction
All cells control when and how much each one of its genes are expressed. This simple statement - one that could be derived simply from observing cellular behavior - brings up many questions that we can begin to lay out using our Design Challenge rubric.
Trying to define "gene expression"
The first thing we need to do, however, is to define what it means when we say that a gene is "expressed". If the gene encodes a protein, one might reasonably propose that "expression" of a gene means how much functional protein is made. But what if the gene does not encode a protein, but rather some functional RNA. Then, in this case, "expressed might mean how much of the functional RNA is made. Yet another person might reasonably suggest that "expression" just refers to the initial step in creating a copy of the genomic information. By that definition, one might want to count how many full-length transcripts are being made. Is it the number of end products encoded by the genomic information or is it the number of reads of the information that is important to properly describe "expression". Unfortunately, in practice we often find that the definition depends on the context of the discussion. Keep that in mind. For the sake of making sure that we are talking about the same thing, in Bis2A we'll try to use the term "expression" primarily to describe the creation of the final functional product(s). Depending on the specific case, the final product may be a protein or RNA species.
The design challenge of regulating gene expression
To drive this discussion from a design challenge perspective, we can formally stipulate that the "big problem" we are interested in understating is that of regulating protein abundance in a cell. Problem: The abundance of each functional protein must be regulated. We can then start by posing subproblems:
Let's take a moment, though, first to reload a couple of ideas. The process of gene expression requires multiple steps depending on what the fate of the final product will be. In the case of structural and regulatory RNAs (i.e. tRNA, rRNA, snRNA, etc.) the process requires that a gene be transcribed and that any needed post-transcriptional processing take place. In the case of a protein coding gene, the transcript must also be translated into protein and if required, modifications to the protein must also be made. Of course, both transcription and translation are multi-step processes and most those sub-steps are also potential sites of control.
Some of the subproblems might therefore be:
1. It is reasonable to postulate that there must be some mechanism(s) to regulate the first step of this multi-step process, the initiation of transcription (just getting things started). So, we could state, "we need a mechanism to regulate the initiation of transcription." We could also turn this into a question and ask, "how can the initiation of transcription be accomplished"?
2. We can use similar thinking to state, "we need a mechanism for regulating the end of transcription" or to ask "how is transcription terminated?"
3. Using this convention we can state, "we need to regulate the initiation of translation and the stop of translation".
4. We've talked only about synthesis of protein and RNA. It is quite reasonable to also state, "we need a mechanisms to regulate the degradation of RNA and protein."
Focusing on transcription
In this course we begin by focusing primarily on examining the first couple of problems/questions, the regulation of transcription initiation and termination - from genomic information to a functional RNA, either ready as is (e.g. in the case of a functional RNA) or ready for translation. This allows us to examine some fundamental concepts regarding the regulation of gene expression and to examine a few real examples of those concepts in action.
Suggested discussion
Why is it important to regulate gene expression why not just express all genes all of the time?
Create a list of hypotheses with your classmates of reasons why the regulation of gene expression is important for bacteria and archaea and for eukaryotes. Instead of bacteria vs. eukaryotes, you may also want to consider contrasting reasons gene regulation is important for unicellular organisms versus multi-cellular organisms or communities of unicellular organisms (like colonies of bacteria).
Subproblems for transcription and the activity or RNA polymerase
Let us consider a protein coding gene and work through some logic. We start by imagining a simple case, where a protein-coding gene is encoded by a single contiguous stretch of DNA. We know that to transcribe this gene an RNA polymerase will need to be recruited to the start of the coding region. The RNA polymerase is not "smart" per se. There needs to be some mechanism, based on chemical logic, to help recruit the RNA polymerase to the start of the protein coding gene. Likewise, if this process is to be regulated, there needs to be some mechanism, or mechanisms, to dictate when an RNA polymerase should be recruited to the start of a gene, when it should not, and/or if it is recruited to the DNA whether or not it should actually begin transcription and how many times this process should happen. Note, that the previous sentence, has several distinct subproblems/questions (e.g. when is the polymerase recruited?; if recruited, should it start transcription?; if it starts transcription, how many times should this process repeat?). We can also reasonably infer, that there will need to be some mechanisms to "instruct" (more anthropomorphisms) the polymerase to stop transcription. Finally, since the role of transcription is to create RNA copies of the genome segments, we should also consider problems/questions related to other factors that influence the abundance of RNA, like mechanisms of degradation. There must be some mechanisms and these will likely be involved in the regulation of this process.
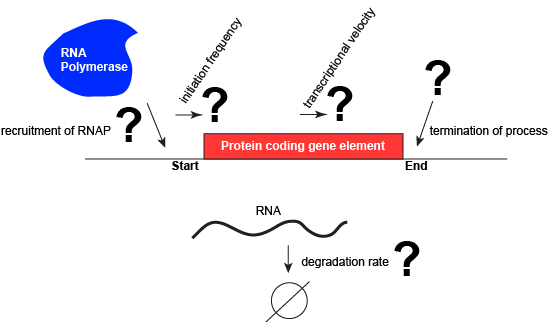
A schematic showing a protein coding gene and some of the questions or problems that we need to ask ourselves or alternatively problems we need to know solutions for if we are to understand how regulation of the transcriptional portion of the gene's expression is regulated.
Attribution: Marc T. Facciotti (own work)
Activation and Repression of Transcription
Some basics
Let us consider a protein coding gene and work through some logic. We start by imagining a simple case, where a protein-coding gene is encoded by a single contiguous stretch of DNA. We know that to transcribe this gene an RNA polymerase will need to be recruited to the start of the coding region. The RNA polymerase is not "smart" per se. There needs to be some mechanism, based on chemical logic, to help recruit the RNA polymerase to the start of the protein coding gene. Likewise, if this process is to be regulated, there needs to be some mechanism, or mechanisms, to dictate when an RNA polymerase should be recruited to the start of a gene, when it should not, and/or if it is recruited to the DNA, whether or not it should actually begin transcription and how many times this process should happen. Note, that the previous sentence, has several distinct subproblems/questions (e.g. when is the polymerase recruited?, if recruited should it start transcription?, if it starts transcription, how many times should this process repeat?). We can also reasonably infer, that there will need to be some mechanisms to "instruct" (more anthropomorphisms) the polymerase to stop transcription.
Recruiting RNA polymerase to specific sites
To initiate transcription, the RNA polymerase must be recruited to a segment of DNA near the start of a region of DNA encoding a functional transcript. The function of the RNA polymerase as described so far, however, is not to bind specific sequences but rather to move along any segment of DNA. Finding a way to recruit the polymerase to a specific site therefore seems contradictory to its usual behavior. Explaining this contradiction requires us to invoke something new. Either transcription can start anywhere and just those events that lead to a full productive transcript do anything useuful or something other than the RNA polymerase itself helps to recruit the enzyme to the beginning of a gene. The latter, we now take for granted, is indeed the case.
The recruitment of the RNA polymerase is mediated by proteins called general transcription factors. In bacteria, they have a special name: sigma factors. In archaea they are called TATA binding protein and transcription factor IIB. In eukaryotes, relatives of the archaeal proteins function in conjunction with numerous others to recruit the RNA polymerase. The general transcription factors have at least two basic functions: (1) They are able to chemically recognize a specific sequence of DNA and (2) they are able to bind the RNA polymerase. Together these two functions of general transcription factors solve the problem of recruiting an enzyme that is otherwise not capable of binding a specific DNA site. In some texts, the general transcription factors (and particularly the sigma factor varieties) are said to be part of the RNA polymerase. While they are certainly part of the complex when they help to target the RNA polymerase they do not continue with the RNA polymerase after it starts transcription.
The DNA site to which an RNA polymerase is recruited has a special name. It is called a promoter. While the DNA sequences of different promoters need not be exactly the same, different promoter sequences typically do have some special chemical properties in common. Obviously, one property is that they are able to associate with an RNA polymerase. In addition, the promoter usually has a DNA sequence that facilitates the dissociation of the double stranded DNA such that the polymerase can begin reading and transcription the coding region. (Note: technically we could have broken down the properties of the promoter into design challenge subproblems. In this case we skipped it, but you should still be able to step backwards and create the problem statements and or relevant questions once you find out about promoters).
In nearly all cases, but particularly in eukaryotic systems the complex of proteins that assembles with the RNA polymerase at promoters (typically called the pre-initiation complex) can number in the tens of proteins. Each of these other proteins has specific function but this is far to too much detail to dive into for Bis2A.
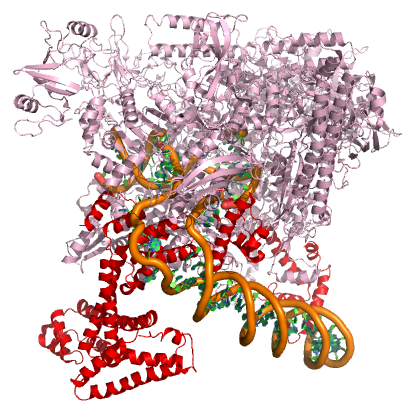
A model of the E. coli pre-initiation complex. The sigma factor is colored red. The DNA is depicted as orange tubes and opposing ring structures. The rest of the pre-initiation complex is colored pink. Note that the DNA has regions of double helix and an open structure inside the PIC.
Attribution: Structure derived from PDB coordinates (4YLN) Marc T. Facciotti (own work)
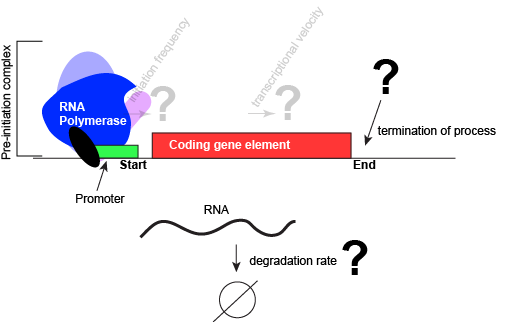
An abstract model of a generic transcriptional unit shown above with the addition of a promoter and PIC. Questions noted earlier that will likely not be covered in Bis2a have been grayed out.
Attribution: Marc T. Facciotti (own work)
Suggested discussion
How does general transcription factor recognize a specific sequence of DNA? What might be the chemical basis for this? How, by contrast might the interaction of a protein or enzyme that interacts with DNA non-specifically differ? Think about functional groups and propose a hypothesis.
States of a regulated promoter
Since promoters recruit an RNA polymerase these sites and the assembly of the pre-initiation complex are obvious sites for regulating the first steps of gene expression. At the level of transcription initiation, we often classify promoter into one of three classes. The first is called constitutive. Constitutive promoters are generally not regulated very strongly. Their base state is "on". When the constitutive transcription from a promoter is very high (relative to most other promoters), we will colloquially call that promoter a "strong constitutive" promoter. By contrast, if the amount of transcription from a constitutive promoter is low (relative to most other promoters) we will call that promoter a "weak constitutive" promoter.
A second way to classify promoters by the use of the term activated or equivalently, induced. These interchangeable terms are used to describe promoters that are sensitive to some external stimulus and respond to said stimulus by increasing transcription. Activated promoters have a base state generally exhibits little to no transcription. Transcription is then "activated" in response to a stimulus - the stimulus turns the promoter "on".
Finally, the third term used to classify promoters is by the use of the term repressed. These promoters also respond to stimuli but do so by decreasing transcription. The base state for these promoters is some basal level of transcription and the stimulus acts to turn down or repress transcription. Transcription is "repressed" in response to a stimulus - the stimulus turns the promoter "off".
The examples given above assumed that a single stimulus acts to regulate promoters. While this is the simplest case, many promoters may integrate different types of information and may be alternately activated by some stimuli and repressed by other stimuli.
Transcription factors help to regulate the behavior of a promoter
How are promoters sensitive to external stimuli? Mechanistically, in both activation and repression, require regulatory proteins to change the transcriptional output of the gene being observed. The proteins responsible for helping to regulate expression are generally called transcription factors. The specific DNA sequences bound by transcription factors are often called operators and in many cases the operators are very close to the promoter sequences.
Here's where the nomenclature gets potentially confusing - particularly when comparing across bacterial and eukaryotic research. In bacterial research, if the transcription factor acts by binding DNA and the RNA polymerase in a way that increases transcription, then it is typically called an activator. If, by contrast, the transcription factor acts by binding DNA to repress or decrease transcription of the gene then it is called a repressor.
Why are the classifications of activator and repressor potentially problematic? These terms, describe in idealized single functions. While this may be true in the case of some transcription factors, in reality other transcription factors may act to activate gene expression in some conditions while repressing in other conditions. Some transcription factors will simply act to modulate expression either up or down depending on context rather than shutting transcription "off" or turning it completely "on". To circumvent some of this possible confusion, some of your instructors prefer to avoid using the terms activator and repressor and instead prefer to simply discuss the activity of transcription various transcription factors as either a positive or a negative influence on gene expression in specific cases. If these terms are used, you might hear your instructor saying that the transcription factor in question ACTS LIKE/AS a repressor or that it ACTS LIKE/AS an activator, taking care not to call it simply an activator or repressor. It is more likely however that you will hear them say that a transcription factor is acting to positively or negatively influence transcription.
CAUTION: Depending on your instructor, you may cover a few real biological examples of positive and negative regulatory mechanisms. These specific examples will use the common names of the transcription factors - since the the examples will typically be drawn from the bacterial literature, the names of the transcription factors may include the terms "repressor" or "activator". These names are relics of when they were first discovered. Try to spend more time examining the logic of how the system works than trying to commit to memory any special properties of that specific protein to all other cases with the same label. That is, just because a protein is labeled as a repressor does not mean that it exclusively acts as a negative regulator in all cases or that it interacts with external signals in the exact same was as the example.
Suggested discussion
What types of interactions do you think happen between the amino acids of the transcription factor and the double helix of the DNA? How do transcription factors recognize their binding site on the DNA?
Allosteric Modulators of Regulatory Proteins
The activity of many proteins, including regulatory proteins and various transcription factors, can be allosterically modulated various factors, including by the relative abundance of small molecules in the cell. These small molecules are often referred to as inducers or co-repressorsor co-activators and are often metabolites, such as lactose or tryptophan or small regulatory molecules, such as cAMP or GTP. These interactions allow the TF to be responsive to environmental conditions and to modulate its function accordingly. It is helping to make a decision about whether to transcribe a gene or not depending on the abundance of the environmental signal.
Let us imagine a negative transcriptional regulator. In the most simple case we've considered so far, transcription of gene with a binding site for this transcription factor would be low when the TF is present and high when the TF is absent. We can now add a small molecule to this model. In this case the small molecule is able to bind the negative transcriptional regulator through sets of complementary hydrogen and ionic bonds. In this first example we will consider the case where the binding of the small molecule to the TF induces a conformational change to the TF that severely reduces its ability to bind DNA. If this is the case, the negative regulator - once bound by its small molecule - would release from the DNA. This would thereby relieve the negative influence and lead to increased transcription. This regulatory logic might be appropriate to have evolved in the following scenario: a small molecule food-stuff is typically absent from the environment. Therefore, genes encoding enzymes that will degrade/use that food should be kept "off" most of the time to preserve the cellular energy that their synthesis would use. This could be accomplished by the action of a negative regulator. When the food-stuff appears in the environment it would be appropriate for the enzymes responsible for its processing to be expressed. The food-stuff could then act by binding to the negative regulator, changing the TF's conformation, causing its release from the DNA and turning on transcription of the processing enzymes.
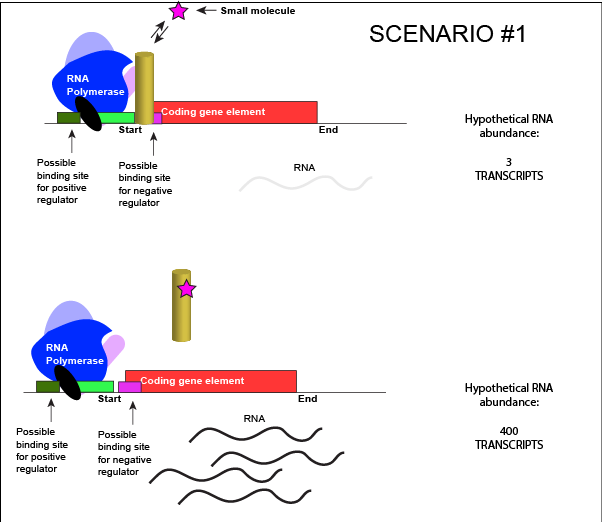
An abstract model of a generic transcriptional unit regulated by a negative regulator whose activity is modulated by a small molecule (depicted by a star). In this case, binding of the small molecule causes the TF to release from the DNA.
Attribution: Marc T. Facciotti (own work)
We can consider a second model for how a negatively acting TF might interact with a small molecule. In this case, the TF alone is unable to bind its regulatory site on the DNA. However, when a small molecule binds to the TF a conformational change occurs that reorients DNA binding amino-acids into the "correct" orientation for DNA binding. The TF-small molecule complex now binds to the DNA and acts to negatively influence transcription.
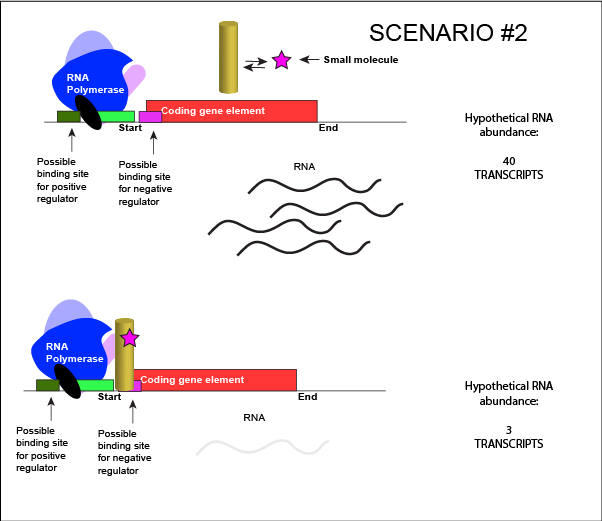
An abstract model of a generic transcriptional unit regulated by a negative regulator whose activity is modulated by a small molecule (depicted by a star). In this case, binding of the small molecule causes the TF to bind to the DNA.
Attribution: Marc T. Facciotti (own work)
Note how the activity of the TF can be modulated in distinctly different ways by a small molecule. Depending on the protein, the binding of this external signal can either cause binding of the TF-small molecule complex to DNA OR binding of the small molecule can cause the release of the TF-small molecule complex from the DNA. The same types of examples can be worked up for a positive regulator.
In both cases proposed above, the binding of a small molecule to a TF will be dependent on how strongly the TF interacts with the small molecule. This will depend on the types and spatial orientation of the protein's chemical functional groups, their protonation states (if applicable), and the complementary functional groups on the small molecule. It should not be surprising, therefore, to learn that the binding of the small molecule to the TF will be dependent on various factors, including but not limited to the relative concentrations of small-molecule and TF and pH.
Is it positive or negative regulation?
Resolving a common point of confusion
At this point, it is not uncommon for many Bis2a students to be slightly confused about how to determine if a transcription factor is acting as a positive or negative regulator. This confusion often comes after a discussion of the possible modes that stimulus (i.e. small molecule) can influence the activity of a transcription factor. This is not too surprising. In the examples above, the binding of a effector molecule to a transcription factor could have one of two different effects: (1) binding of the effector molecule could induce a DNA-bound transcription factor to release from its binding site, derepressing a promoter, and "turning on" gene expression. (2) binding of the effector molecule to the transcription factor could induce the TF to bind to its DNA binding site, repressing transcription and "turning off" gene expression. In the first case it might appear that the TF is acting to positively regulate expression, while in the second example it might appear that the TF is acting negatively.
However, in both examples above, the TF is acting as a negative regulator. To determine this we look at what happens when the TF binds DNA (whether a small molecule is bound to the TF or not). In both cases, binding of the TF to DNA represses transcription. The TF is therefore acting as a negative regulator. A similar analysis can be done with positively acting TFs.
Note that in some cases a TF may act as a positive regulator at one promoter and negative regulator at a different promoter so describing the behavior of the TF on a per case basis is often important (reading too much from the name it has been assigned can be misleading sometimes). Other TF protein can act alternately as both positive or negative regulators of the same promoter depending on conditions. Again, describing the behavior of the TF specifically for each case is advised.
A genetic test for positive or negative regulatory function of a TF
How does one determine if a regulatory protein functions in a positive or negative way? A simple genetic test is to ask "what happens to expression if the regulatory protein is absent?" This can be accomplished by removing the coding gene for the transcription factor from the genome. If a transcription factor acts positively, then its presence is required to activate transcription. In its absence, there is no regulatory protein, therefore no activation, and the outcome is lower transcription levels of a target gene. The opposite is true for a transcription factor acting negatively. In its absence expression should be increased, because the gene keeping expression low is no longer around.
Termination of Transcription and RNA degradation
Termination of transcription in bacteria
Once a gene is transcribed, the bacterial polymerase needs to be instructed to dissociate from the DNA template and liberate the newly made mRNA. Depending on the gene being transcribed, there are two kinds of termination signals. One is protein-based and the other is RNA-based. Rho-dependent termination is controlled by the rho protein, which tracks along behind the polymerase on the growing mRNA chain. Near the end of the gene, the polymerase encounters a run of G nucleotides on the DNA template and it stalls. As a result, the rho protein collides with the polymerase. The interaction with rho releases the mRNA from the transcription bubble.
Rho-independent termination is controlled by specific sequences in the DNA template strand. As the polymerase nears the end of the gene being transcribed, it encounters a region rich in C–G nucleotides. The mRNA folds back on itself, and the complementary C–G nucleotides bind together. The result is a stable hairpin that causes the polymerase to stall as soon as it begins to transcribe a region rich in A–T nucleotides. The complementary U–A region of the mRNA transcript forms only a weak interaction with the template DNA. This, coupled with the stalled polymerase, induces enough instability for the core enzyme to break away and liberate the new mRNA transcript.
Termination of transcription in eukaryotes
The termination of transcription is different for the different polymerases. Unlike in prokaryotes, elongation by RNA polymerase II in eukaryotes takes place 1,000–2,000 nucleotides beyond the end of the gene being transcribed. This pre-mRNA tail is subsequently removed by cleavage during mRNA processing. On the other hand, RNA polymerases I and III require termination signals. Genes transcribed by RNA polymerase I contain a specific 18-nucleotide sequence that is recognized by a termination protein. The process of termination in RNA polymerase III involves an mRNA hairpin similar to rho-independent termination of transcription in prokaryotes.
Termination of transcription in archaea
Termination of transcription in the archaea is far less studied than in the other two domains of life and is still not well understood. While the functional details are likely to resemble mechanisms that have been seen in the other domains of life the details are beyond the scope of this course.
Degradation of RNA
The lifetimes of different RNA species in the cell can vary dramatically, from seconds to hours. The mean lifetime of mRNA can also vary dramatically depending on the organism. For instance, the median lifetime for mRNA in E. coli is ~5 minutes. The half-life of mRNA in yeast is ~20 minutes and 600 minutes for human cells. Some of the degradation is "targeted". That is, some transcripts include a short sequence that targets them for RNA degrading enzymes, speeding the degradation rate. It doesn't take too much imagination to infer that this process might also be evolutionarily tuned for different genes. Simply realizing that degradation - and the tuning of degradation - can also be a factor in controlling the expression of a gene is sufficient for Bis2a.
Tuning termination
Like all other biological processes, the termination of transcription is not perfect. Sometimes, the RNA polymerase is able to read through terminator sequences and transcribe adjacent genes. This is particularly true in bacterial and archaeal genomes where the density of coding genes is high. This transcriptional read-through can have various effects but the most common two outcomes are: (1) If the adjacent genes are encoded on the same DNA strand as the actively transcribed gene, the adjacent genes may also be transcribed. (2) If the adjacent genes are transcribed on the opposite strand of the actively transcribed genes, RNA polymerase read-through by interfere with polymerases actively transcribing the neighboring gene. Not surprisingly, biology has in some instances evolved mechanisms that both act to minimize the influence of read-through and to take advantage of it. Therefore, the "strength" of a terminator - and its effectiveness of terminating transcription in a particular direction - are "tuned" by Nature and "used" (note the anthropomorphism in quotes) to regulate the expression of genes.
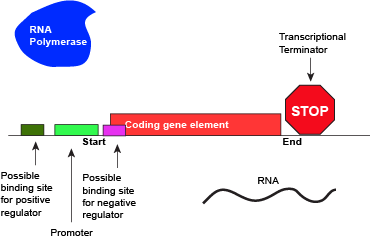
An abstract model of a full basic transcriptional unit and the various "parts" encoded on the DNA that may influence the expression of the gene. We expect students in Bis2A to be able to recreate a similar conceptual figure from memory.
Attribution: Marc T. Facciotti (own work)
Summary of gene regulation
In the preceding text we have examined several ways to start solving some of the design challenges associated with regulating the amount of transcript that is created for a single coding region of the genome. We have looked in abstract terms at some of the processes responsible for controlling the initiation of transcription, how these may be made sensitive to environmental factors, and very briefly at the processes that terminate transcription and handle the active degradation of RNA. We have avoided more Each of these processes can be quantitatively tuned by nature to be "stronger" or "weaker". It is important to realize that the real values of "strength" (e.g. promoter strength, degradation rates, etc.) influence the behavior of the overall process in potentially functionally important ways.
Examples of Bacterial Gene Regulation
This section describes two examples of transcriptional regulation in bacteria. These are presented as illustrative examples. Use these examples to learn some basic principles about mechanisms of transcriptional regulation. Be on the lookout in class, in discussion, and in the study-guides for extensions of these ideas and use these to explain the regulatory mechanisms used for regulating other genes.
Gene Regulation Examples in E. coli
The DNA of bacteria and archaea are usually organized into one or more circular chromosomes in the cytoplasm. The dense aggregate of DNA that can be seen in electron micrographs is called the nucleoid. In bacteria and archaea, genes, whose expression needs to be tightly coordinated (e.g. genes encoding proteins that are involved in the same biochemical pathway) are often grouped closely together in the genome. When the expression of multiple genes is controlled by the same promoter and a single transcript is produced these expression units are called operons. For example, in the bacterium Escherschia coli all of the genes needed to utilize lactose are encoded next to one another in the genome. This arrangement is called the lactose (or lac) operon. It is often the case in bacteria and archaea that nearly 50% of all genes are encoded into operons of two or more genes.
The Role of the Promoter
The first level of control of gene expression is at the promoter itself. Some promoters recruit RNA polymerase and turn those DNA-protein binding events into transcripts more efficiently than other promoters. This intrinsic property of a promoter, it's ability to produce transcript at a particular rate, is referred to as promoter strength. The stronger the promoter, the more RNA is made in any given time period. Promoter strength can be "tuned" by Nature in very small or very large steps by changing the nucleotide sequence the promoter (e.g. mutating the promoter). This results in families of promoters with different strengths that can be used to control the maximum rate of gene expression for certain genes.
UC Davis Undergraduate Connection:
A group of UC Davis students interested in synthetic biology used this idea to create synthetic promoter libraries for engineering microbes as part of their design project for the 2011 iGEM competition.
Example #1: Trp Operon
Logic for regulating tryptophan biosynthesis
E. coli, like all organisms, needs to either synthesize or consume amino acids to survive. The amino acid tryptophan is one such amino acid. E. colican either import tryptophan from the environment (eating what it can scavenge from the world around it) or synthesize tryptophan de novo using enzymes that are encoded by five genes. These five genes are encoded next to each other in the E. coli genome into what is called the tryptophan (trp) operon (Figure below). If tryptophan is present in the environment, then E. coli does not need to synthesize it and the switch controlling the activation of the genes in the trp operon is switched off. However, when environmental tryptophan availability is low, the switch controlling the operon is turned on, transcription is initiated, the genes are expressed, and tryptophan is synthesized. See the figure and paragraphs below for a mechanistic explanation.
Organization of the trp operon
Five genomic regions encoding tryptophan biosynthesis enzymes are arranged sequentially on the chromosome and are under the control of a single promoter - they are organized into an operon. Just before the coding region is the transcriptional start site. This is, as the name implies, the location where the RNA polymerase starts a new transcript. The promoter sequence is further upstream of the transcriptional start site.
A DNA sequence called an "operator" is also encoded between the promoter and the first trp coding gene. This operator is the DNA sequence to which the transcription factor protein will bind.
A few more details regarding TF binding sites
It should be noted that the use of the term "operator" is limited to just a few regulatory systems and almost always refers to the binding site for a negatively acting transcription factor. Conceptually what you need to remember is that there are sites on the DNA that interact with regulatory proteins allowing them to perform their appropriate function (e.g. repress or activate transcription). This theme will be repeated universally across biology whether the "operator" term is used or not.
Moreover, while the specific examples you will be show depict TF binding sites in their known locations, these locations are not universal to all systems. Transcription factor binding sites can vary in location relative to the promoter. There are some patterns (e.g. positive regulators are often upstream of the promoter and negative regulators bind downstream), but these generalizations are not true for all cases. Again, the key thing to remember is that transcription factors (both positive and negatively acting) have binding sites with which they interact to help regulate the initiation of transcription by RNA polymerase.
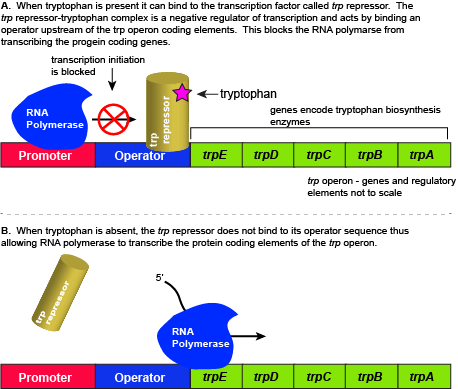
The five genes that are needed to synthesize tryptophan in E. coli are located next to each other in the trp operon. When tryptophan is plentiful, two tryptophan molecules bind to the transcription factor and allow the TF-tryptophan complex to bind at the operator sequence. This physically blocks the RNA polymerase from transcribing the tryptophan biosynthesis genes. When tryptophan is absent, the transcription factor does not bind to the operator and the genes are transcribed.
Attribution: Marc T. Facciotti (own work)
Regulation of the trp operon
When tryptophan is present in the cell: two tryptophan molecules bind to the trp repressor protein. When tryptophan binds to the transcription factor it causes a conformational change in the protein which now allows the TF-tryptophan complex to bind to the trp operator sequence. Binding of the tryptophan–repressor complex at the operator physically prevents the RNA polymerase from binding, and transcribing the downstream genes. When tryptophan is not present in the cell, the transcription factor does not bind to the operator; therefore, the transcription proceeds, the tryptophan utilization genes are transcribed and translated, and tryptophan is thus synthesized.
Since the transcription factor actively binds to the operator to keep the genes turned off, the trp operon is said to be "negatively regulated" and the proteins that bind to the operator to silence trp expression are negative regulators.
Suggested discussion
Do you think that the constitutive expression levels of the trp operon are high or low? Why?
Suggestion discussion
Suppose nature took a different approach to regulating the trp operon. Design a method for regulating the expression of the trp operon with a positive regulator instead of a negative regulator. (hint: we ask this kind of question all of the time on exams)
External link
Watch this video to learn more about the trp operon.
Example #2: The lac operon
Rationale for studying the lac operon
In this example, we examine the regulation of genes encoding proteins whose physiological role is to import and assimilate the disaccharide lactose, the lac operon. The story of the regulation of lac operon is a common example used in many introductory biology classes to illustrate basic principles of inducible gene regulation. We choose to describe this example second because it is, in our estimation, more complicated than the previous example involving the activity of a single negatively acting transcription factor. By contrast, the regulation of the lac operon is, in our opinion, a wonderful example of how the coordinated activity of both positive and negative regulators around the same promoter can be used to integrate multiple different sources of cellular information to regulate the expression of genes.
As you go through this example, keep in mind the last point. For many Bis2a instructors it is more important for you to learn the lac operon story than it is to know the logic table presented below. For those instructors for which this is the case, they will usually make a point to let you know and often deliberately not include exam questions about the lac operon. Rather they will test you on whether you understood the basis underlying the regulatory mechanisms that you study. If it's not clear what the instructor wants you should ask.
The utilization of lactose
Lactose is a disaccharide composed of the hexoses glucose and galactose. It is commonly found in high abundance in milk and some milk products. As one can imagine, the disaccharide can be an important food-stuff for microbes that are able to utilize its two hexoses. E. coli is able to use multiple different sugars as energy and carbon sources, including lactose and the lac operon is a structure that encodes the genes necessary to acquire and process lactose from the local environment. Lactose, however, has not been frequently encountered by E. coli during its evolution and therefore the genes of the lac operon must typically be repressed (i.e. "turned off") when lactose is absent. Driving transcription of these genes when lactose is absent would waste precious cellular energy. By contrast, when lactose is present, it would make logical sense for the genes responsible for the utilization of the sugar to be expressed (i.e. "turned on"). So far the story is very similar to that of the tryptophan operon described above.
However, there is a catch. Experiments conducted in the 1950's by Jacob and Monod clearly demonstrated that E. coli prefers to utilize all the glucose present in the environment before it begins to utilize lactose. This means that the mechanism used to decide whether or not to express the lactose utilization genes must be able to integrate two types of information (1) the concentration of glucose and (2) the concentration of lactose. While this could theoretically be accomplished in multiple ways, we will examine how the lac operon accomplishes this by using multiple transcription factors.
The transcriptional regulators of the lac operon
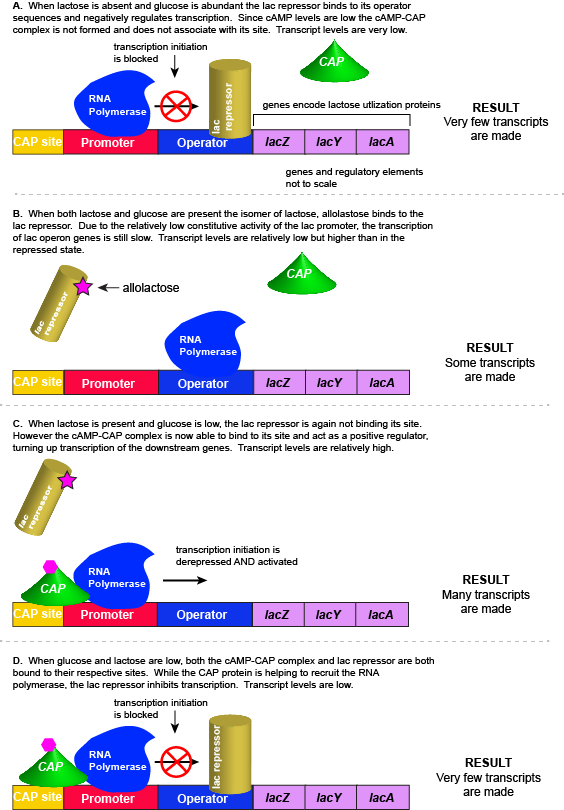
The lac repressor - a direct sensor of lactose
As noted, the lac operon normally has very low to no transcriptional output in the absence of lactose. This is due to two factors: (1) the constitutive promoter strength for the operon is relatively low and (2) the constant presence of the LacI repressor protein negatively influences transcription. This protein binds to the operator site near the promoter and blocks RNA polymerase from transcribing the lac operon genes. By contrast, if lactose is present, lactose will bind to the LacI protein, inducing a conformational change that prevents LacI-lactose complex from binding to its binding sites. Therefore, when lactose is present the negative regulatory LacI is not bound to the its binding site and transcription of lactose utilizing genes can proceed.
CAP protein - an indirect sensor of glucose
In E. coli, when glucose levels drop, the small molecule cyclic AMP (cAMP) begins to accumulate in the cell. cAMP is a common signaling molecule that is involved in glucose and energy metabolism in many organisms. When glucose levels decline in the cell, the increasing concentrations of cAMP allow this compound to bind to the positive transcriptional regulator called catabolite activator protein (CAP) - also referred to as CRP. cAMP-CAP complex has many sites located throughout the E. coli genome and many of these sites are located near the promoters of many operons that control the processing of various sugars.
In the lac operon, the cAMP-CAP binding site is located upstream of the promoter. Binding of cAMP-CAP to the DNA helps to recruit and retain RNA polymerase to the promoter. The increased occupancy of RNA polymerase to its promoter, in turn, results in increased transcriptional output. In this case the CAP protein is acting as a positive regulator.
Note that the CAP-cAMP complex can, in other operons, also act as a negative regulator depending upon where the binding site for CAP-cAMP complex is located relative to the RNA polymerase binding site.
Putting it all together: Inducing expression of the lac operon
For the lac operon to be activated, two conditions must be met. First, the level of glucose must be very low or non-existent. Second, lactose must be present. Only when glucose is absent and lactose is present will the lac operon be transcribed. When this condition is achieved the LacI-lactose complex dissociates the negative regulator from near the promoter, freeing the RNA polymerase to transcribe the operon's genes. Moreover, high cAMP (indirectly indicative of low glucose) levels trigger the formation of the CAP-cAMP complex. This TF-inducer pair now bind near the promoter and act to positively recruit the RNA polymerase. This added positive influence boosts transcriptional output and lactose can be efficiently utilized. The mechanistic output of other combinations of binary glucose and lactose conditions are descried in the table below and in the figure that follows.
A more nuanced view of lac repressor function
The description of the lac repressor's function correctly describes the logic of the control mechanism used around the lac promoter. However, the molecular description of binding sites is a bit overly simplified. In reality the lac repressor has three similar, but not identical, binding sites called Operator 1, Operator 2, and Operator 3. Operator 1 is very close to the transcript start site (denoted +1). Operator 2 is located about +400nt into the coding region of the LacZ protein. Operator 3 is located about -80nt before the transcript start site (just "outside" of the CAP binding site).
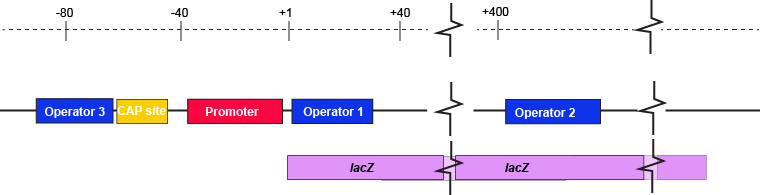
The lac operon regulatory region depicting the promoter, three lac operators, and CAP binding site. The coding region for the Lac Z protein is also shown relative to the operator sequences. Note that two of the operators are in the protein coding region - there are multiple different types of information simultaneously encoded in the DNA.
Attribution: Marc T. Facciotti (own work)
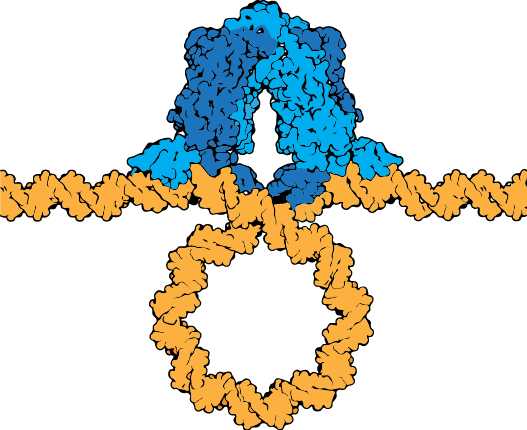
The lac repressor tetramer (blue) depicted binding two operators on a strand of looped DNA (orange).
Attribution: Marc T. Facciotti (own work) - Adapted from Goodsell (https://pdb101.rcsb.org/motm/39)