Winter_2023_Bis2A_Facciotti_Reading_01
- Page ID
- 102209
Learning Objectives Associated with Winter_2023_Bis2a_Facciotti_Reading_01
|
Introduction to Biomolecules in BIS2A
Some context and motivation
In BIS2A we want to learn how living systems "work". To accomplish this we need to understand the "stuff" biological systems. We want to know what "stuff" organisms are made of and how the properties of this "stuff" contribute to organismal forms and behaviors. This requires us to dig into a little chemistry — the science of the "stuff" that makes up the world we know. By doing so, we can show that many of the biological processes we care about arise directly from the chemical properties of the "stuff" that makes up living systems. Developing a functional understanding of some basic chemical concepts can, therefore, be tremendously useful in thinking about how to solve problems in medicine, energy, and environment by attacking them at their molecular levels. For these reasons, we begin a study of biomolecules and their chemistry.
Importance of chemical composition
In this course, we want you to learn how to classify major classes of biological macromolecules into groups by looking at their chemical composition and, based on this composition, to also infer some properties they might have. For example, you will learn to recognize the molecular structures of carbohydrates. You'll learn to do so by spotting chemical "parts" on carbohydrates called hydroxyl groups and appreciate that those chemical "parts" can form special types of interactions called "hydrogen bonds". You'll discover how hydrogen bonds give carbohydrates special properties that govern how they interact with water and other molecules and how these interactions give different properties to carbohydrates like simple sugars and starches. This will shed light on the functional roles these molecules play in living systems, how they are built, and how they can be broken down. We will also look at other types of common macromolecules in biology in the same way.
Linking molecular structure to function
In biology, structure is often said to "encode" function. That is, the shape and physical properties of something governs what it can do. In this course we want to make this general idea a bit more specific for you and begin to help you form the conceptual ideas that will help you explain how the chemical structure and properties of biomolecules can contribute to their functions.
For example, biological membranes have the biological function of defining the boundaries between the inside and outside of a cell or compartments within a cell. The membranes also serve as selective barriers that allow certain things to pass into and out of a cell while excluding others substances from crossing the barrier. These are very important functions indeed! The chemical structures of membranes determine these functional properties. You will learn, for instance, that most membranes are composed of two layers of molecules called phospholipids and that phospholipids can be schematically broken down into two chemically distinct subregions, a so-called hydrophilic head group and a hydrophobic tail group. Each of these chemical subregions and the chemical "parts" they are made of, contribute directly to determining the structure and permeability of cell membranes.
Likewise, we will also learn about important biological molecules called enzymes. Enzymes help catalyze many of the chemical reactions in an organism, including how they process the food they eat, how an organism senses and interacts with its environment, and how it replicates (among many other functions). We will learn that the three dimensional structures of enzymes encode their functions and that their structures are determined to a large degree by the number, type, and order of chemicals called amino acids. The properties of these amino acids (and thus how they behave in an enzyme) are directly related to their own chemical "parts".
In sum, while structure "encodes" biological function, structure is itself usually determined by chemistry. Knowing a little bit of chemistry can be powerful in biology.
Objectives and strategy
On these notes we set out to learn a little bit of useful chemistry for biology. Our objectives for the next few weeks are fairly straightforward. First, we want to learn some basic vocabulary, symbols and models that help us communicate about chemistry in biology. Second, we want to learn a few basic ideas about what the "stuff-of-life" is made of and some basic properties of that stuff that help us understand biological phenomena. Third, we want to learn to recognize some of the most common chemical structures in biology and something about their properties. Fourth, we want to understand how some of the chemistries of the "stuff-of-life" can be altered and how this might change the function of a biomolecule. Fifth, we want to develop a vocabulary and learn key ideas that help us talk about how things change at the molecular level in biology.
If you've already taken some chemistry, some of the ideas will be familiar to you. If you haven't taken chemistry - or it's been a long time since you did - don't worry. We won't cover these topics in nearly as theoretically or in as much detail as your chemistry instructor will. We want to extract the most important ideas/concepts that help us function as general biologists. The core ideas and concepts aren't too complicated. However, there will likely be a lot of new vocabulary to learn and a good amount of practice required to get comfortable with thinking about things at the molecular scale. Success comes from practice. So, practice early and often. Strive to learn the new vocabulary quickly and take advantage of practice opportunities to think about molecules. That'll make it much easier to then think about and add new concepts as we go into the next few weeks.
The Periodic Table
The periodic table is a tool for organizing information about the different elements found in nature. Devised by Russian chemist Dmitri Mendeleev (1834–1907) in 1869, the table groups elements that, because of some commonalities of their atomic structure, share certain chemical properties. The atomic structure of elements is responsible for their physical properties including whether they exist as gases, solids, or liquids under specific conditions and their chemical reactivity, a term that refers to their ability to combine and to bond chemically with each other and other elements.
In the periodic table, shown below, the elements are organized and displayed according to their atomic number and are arranged in a series of rows and columns based on shared chemical and physical properties. Besides providing the atomic number for each element, the periodic table also displays the element’s atomic mass. Looking at carbon, for example, its symbol (C) and name appear, and its atomic number of six (in the upper right-hand corner showing the number of protons in the neutral nucleus) and its atomic mass of 12.11 (sum of the mass of electrons, protons, and neutrons).
Hydrogen Bonds
When hydrogen forms a polar covalent bond with an atom of higher electronegativity, the region around the hydrogen will have a fractional positive charge (termed δ+). When this fractional positive charge encounters a partial negative charge (termed δ-) from another electronegative atom to which the hydrogen is NOT bound, AND it is presented to that negative charge in a suitable orientation, a special kind of interaction called a hydrogen bond can form. While chemists are still debating the exact nature of the hydrogen bond, in BIS2A, we like to conceive of it as a weak electrostatic interaction between the δ+ of the hydrogen and the δ- charge on an electronegative atom. We call the molecule that contributes the partially charged hydrogen atom the "hydrogen bond donor" and the atom with the partial negative charge the "hydrogen bond acceptor." We will ask you to learn to recognize common biological hydrogen bond donors and acceptors and to identify putative hydrogen bonds from models of molecular structures.
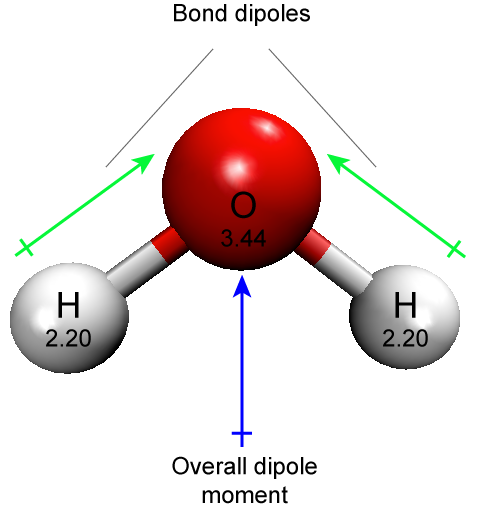
Hydrogen bonds are common in biology both within and between many biomolecules. Hydrogen bonds are also critical interactions between biomolecules and their solvent, water. It is common, as seen in the figure below, to represent hydrogen bonds in figures with dashed lines.
Figure 1: Two water molecules are depicted forming a hydrogen bond (drawn as a dashed blue line). The water molecule on top "donates" a partially charged hydrogen while the water molecule on the bottom accepts that partial charge by presenting a complementary negatively charged oxygen atom. Attribution: Marc T. Facciotti (original work)
Dipoles, Van der Waals Forces, and Pi Interactions
In addition to ionic and hydrogen bonds, there are several other types of non-covalent molecular interactions that we encounter in General Biology. Key among these are dipole-dipole interactions, Van der Waals forces and pi interactions. In this section we briefly describe each of these interactions and some of their underlying basis. Developing a deep and comprehensive theoretical understanding of these interaction types requires a dive into more advanced chemistry. We don’t do that. Rather, we try to provide a more descriptive understanding of these phenomena that will hopefully be useful for interpreting common molecular interactions in biology. Recall that with respect to chemistry, our goals in General Biology are relatively modest. We want students to recognize different chemical interactions between biomolecules, to appreciate that these interactions arise from the unique chemical properties of the elements that make up the molecules, and to appreciate how environmental and chemical factors can change these interactions. If you can identify obvious biological scenarios in which different interactions can take place, you’re doing great!
Dipole-Dipole Interactions
Dipole-dipole interactions are, as the name suggests, simply interactions between two dipoles. Recall how the differences in electronegativities between elements can explain the creation of polar covalent bonds. We describe these polar covalent bonds as permanent dipoles, “hard-coded” by the properties of the elements bonded together. The dipole-dipole interaction is an interaction between two permanent dipoles. If partial charges carrying the same (+ or -) sign interact (i.e. positive interacts with positive or negative interacts with negative) we say that the interacting molecules experience a repulsive dipole-dipole force which pushes the molecules away from one another. If partial charges of opposite sign (i.e. positive interacts with negative) interact, we say that the interacting molecules experience an attractive dipole-dipole force which attracts the molecules to one another.

Figure \(\PageIndex{1}\): Dipole-dipole interactions. Permanent dipoles established through the covalent interaction between atoms A and B can interact via dipole-dipole interactions. Atom B is more electronegative than atom A and thus recruits electrons near it causing an imbalance in charge around the molecule (this is depicted by an oblong electron cloud around nuclei for atoms A and B) - more negative is red; more positive is blue. The imbalance of charge creates a dipole with partial negative charges (delta-) and partial positive charges (delta+). The figure depicts various ways in which two of these dipoles can interact with one another that, depending on orientation, lead to either attractive or repulsive dipole-dipole interactions.
Attribution: Marc T. Facciotti (original work)
The core idea underlying the formation of dipole-dipole interactions - the interaction between two permanent dipoles - should sound familiar. Recall that we describe a hydrogen bond as an electrostatic interaction between a partially charged hydrogen (the positive end of a bond dipole) with a partial negative charge from the negative end of a different bond dipole. While the hydrogen bond has some special properties not discussed in this text, you can think of it as a sub-type of attractive dipole-dipole interaction.
For a deeper dive into dipole-dipole interactions, see this LibreText Chemistry reading.
Van der Waals Forces
All molecules can experience Van der Waals forces, a type of molecular interaction found when molecules get very close together, typically at distances between 4-5 Angstroms. Just for reference, recall that 1 Angstrom = 10-10 meters. Van der Waals forces are, yet again, based on the attraction or repulsion of electrical poles. However, unlike the dipole-dipole interactions discussed above that arise from the interaction between permanent dipoles in molecules, the Van der Waals forces arise from the spontaneous and/or induced transient polarization of molecules. The local polarization (i.e. polarization on a part of a molecule) may last only a short time as electrons dynamically redistribute. When two molecules are close together one of them may spontaneously form a transient dipole (or more accurately, multipole). In response, the second molecule may “sense” the partial charge nearby and react by adjusting its own charge distribution in response, thus becoming polar itself. We say that the first dipole/multipole induces the formation of the second. If the two molecules remain between 4 and 5 Angstroms long enough, this process can repeat, and even synchronize, leading to molecular attraction at very short distances. At distances closer than ~4 Angstroms, electron clouds can overlap and this creates repulsive interaction. While all molecules can engage in Van der Waals interactions, in introductory biology we usually introduce students to these interactions in a discussion of lipid membrane structures. As you will soon discover, Nature creates biological membranes by packing many lipid molecules together at distances that allow many simultaneous Van der Waals interactions to occur. Collectively, these many small and transient interactions between lipid molecules contribute to the stability of membrane structures.

Figure \(\PageIndex{1}\): Schematic of dynamic, induced dipoles involved in Van der Waals forces. Molecules composed of atoms A are depicted approaching one another near the top of the figure. The dynamics (change in time) of these molecules is depicted row-by-row as they change in through arbitrary jumps in time. This dynamic nature is critical to the formation of Van der Waals interactions. Attribution: Marc T. Facciotti (original work)
Pi Interactions
Pi interactions are a type of molecular interaction that biologists typically encounter in when discussing stabilizing interactions in nucleic acid and protein structures. In a course of General Biology, you may also encounter pi interactions in a discussion of protein-DNA interactions. These types of interactions derive their name from the involvement of pi bonds, a specific type of covalent bond between two atoms in which neighboring electron orbitals are close enough to overlap. We’ll leave the underlying discussion of molecular orbital theory for your chemistry course and just say that we usually associate pi bonds with double or triple covalent bonds. In biology, these types of bonds occur in many kinds of molecules, particularly those with so called conjugated pi systems including aromatic ring structures like those seen in some amino acids, vitamins and cofactors, and nucleic acids.

Figure \(\PageIndex{1}\): Examples of molecules with conjugated pi systems. Each of the biomolecules has at least a portion of it that contains so called conjugated pi bonds that can engage in pi interactions with other molecules. With the exception of the retinol molecules the conjugated pi systems are present in planar ring structures. The retinol's conjugated pi system is in the linear hyrodrocarbon portion of the molecule. Arrows point to examples of double bonds. Yellow highlights the systems of delocalized electrons. Attribution: Marc T. Facciotti (original work)
The distribution of electrons within these pi systems can create regions of more negative and more positive charge and thus creating areas that may “attract” or “repel” other charged or partially charged molecules depending on their relative alignments to one another. Again, we can reserve a deeper discussion of pi systems for your upper division chemistry classes. For now, simply appreciate that - once again - the unique properties of the elements that make up molecules contribute to how electrons distribute within those molecules. The often uneven distributions of electrons about the molecule can create local positive and negative charges and when these regions of partial charges on different molecules (or different parts of molecules) come together in appropriate orientations, that electrostatic interactions (attractive and repulsive) can happen.
A TAKE-HOME POINT ON MOLECULAR INTERACTIONS
Hopefully, you appreciate a common theme to our discussion of non-covalent molecular interactions. Whether we consider ionic bonds, hydrogen bonds, dipole-dipole interactions, Van der Waals forces, or pi interactions, all share the feature of being interactions between full or partial electrostatic charges. The key differences between each of these types of interactions types have to do with how the charges arise on molecules (i.e. the atomic basis for the charge) and/or how the charges interact. This depends, of course, on the underlying unique chemical properties of each element and how they behave at the subatomic level with one another - a suitable topic for discussion in your chemistry class.
PRACTICE POST-GUIDE
General Practice
Why: One of the major challenges facing students of General Biology is that we talk about and study things, like cells and molecules, that we aren’t used to seeing in our everyday lives. Molecules are small compared to what we usually deal with. Most of us, even the more experienced learners, can’t easily conjure up a mental image of what a molecule looks like. Even reading stick figure representations of molecules and dealing with the different ways people draw them takes some serious mental energy. To understand and communicate about biology at the molecular scale, however, we need to get better at thinking about, reading, and writing molecular structures. So, let’s practice getting used to reading and writing models of biomolecules.
What to do for practice: Take advantage of the internet! Open a web browser and do a Google search for “biomolecule”. Find at least ten images of chemical structures that are drawn like those we have seen in class (click the image tab in the search results).
Now make your brain do some active practice! Redraw/copy their chemical structures in your sketchpad. Try to find different examples of biomolecules with with the same name but that are drawn differently (e.g. some with the shortcut of implying carbons where two or more lines meet, etc.). Start getting used to the look of these models. We will be using different representations interchangeably in class and likely in your other classes, so we want to become proficient at “reading” them. You want your brain focusing on the key lesson and not on reading the stick figures.
This is good practice for LOs: GC.12 Recognize the symbols and names of the six core elements in biomolecules: C, H, N, O, P, S. GC.13 Interpret (identity elements and bond types) chemical and structural formulas (including 2- and 3-dimensional, condensed, and line-angle) of biomolecules. GC.3 Identify ionic, covalent, polar covalent bonds, hydrogen bonds, and Van der Waals interactions in different types of molecular models.
Why: This course is centered around learning general principles that apply to all living systems - whether you, a furry pet, or some microbe living at the bottom of the sea. As we talk about specific topics it is useful to create a mental scaffold or some other logical structure on which to place our new learnings (a picture that we’re going to fill in over the quarter). One of those common mental structures is the concept a cell. A second, related concept, is relatedness/connectedness of all living systems, and building a mental scaffold on which to place ideas/concepts that are common to all living systems.
How to practice: Revisit your list of properties shared by all living things that you derived in class. Are there ideas that came up in class that are missing from your list? Are there any that you disagree with? Try to really think about this and refine your conclusion. Next, based on our discussion in lecture today, try (re)drawing a conceptual figure for how we might structure a view of a cell from these high-level concepts. Is this figure any different from the figure you drew in class? Force yourself to be a bit more abstract if you didn’t do it the first time so that you make sure to include the “high-level” concepts.
This is good practice for LO: CI.15 Create a conceptual drawing of a cell that reflects how cells manifest and solve several requirements for life. The drawing should include cellular structures and possible interactions with the environment.
PRACTICE EXAM QUESTIONS
Question Q1.1 An easy question—anything like this would likely appear as part of a broader question on an exam. It’s mostly to practice GC.2 Identify the locations of the three core atomic elements (electrons, neutrons, and protons) from a basic model for an atom and describe their basic properties.
Q1.1 A Ca2+ ion has a charge of +2. What can be said about the relative number of subatomic particles in this ion?
- The ion has equal numbers of protons, neutrons, and electrons.
- The ion has two fewer neutrons than protons or electrons.
- The ion has two more protons than neutrons and electrons.
- The ion has two fewer electrons than protons and neutrons.
- There is not sufficient information to reach a conclusion.
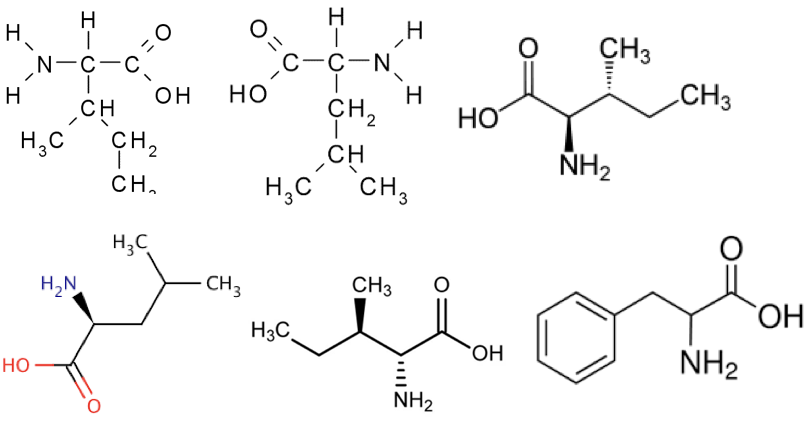
Question Q1.2
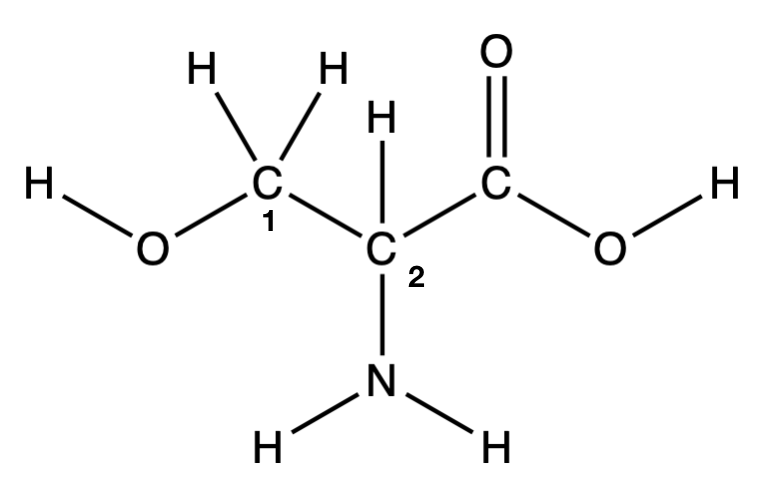
Q1.2: How many unique molecules are represented in the figures below (they are all amino acids—we’ll get very comfortable with these this quarter)?
A. 1
B. 2
C. 3
D. 4
E. 6
Question Q1.3
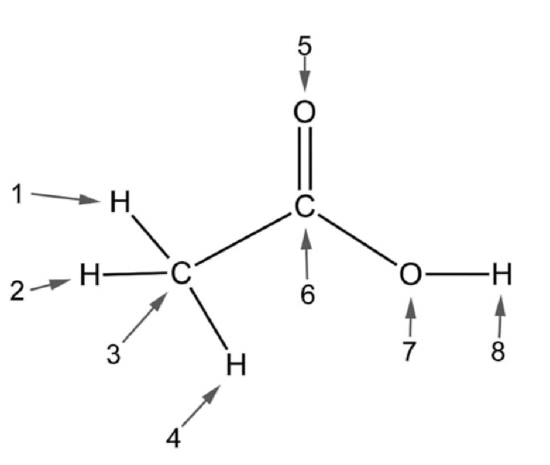
Q1.3: Look at the figure below. Select the answer choice that contains all atoms that can participate in hydrogen bonding with water and no atoms that can’t.
A. 1,2,4,5,7,8
B. 5,7,8
C. 1,2,4,7,8
D. 5,7
E. 7,8