S2024_Bis2a_Namekawa_Regulation_of_Gene_Expression_I
- Page ID
- 132382
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
Introduction to gene regulation
Regulation is all about decision making. Gene regulation is, therefore, all about understanding how cells decide about which genes to turn on, turn off or tune up or tune down. In the following section, we discuss some fundamental mechanisms and principles used by cells to regulate gene expression in response to changes in cellular or external factors. This biology is important for understanding how cells adjust changing environments, including how some cells, in multicellular organisms, become specialized for certain functions (e.g. tissues).
Since the subject of regulation is both a deep and broad topic of study in biology, in Bis2a we don't cover every detail - there are way too many. Rather, as we have done for all other topics, we try to focus on (a) outlining some core logical constructs and questions that you must have when you approach ANY scenario involving regulation, (b) learning some common vocabulary and ubiquitous mechanisms and (c) examining a few concrete examples that illustrate the points made in a and b.
Gene Expression
Introduction
All cells control when and how much each one of its genes is expressed. This simple statement - one that could be derived from observing cellular behavior - brings up many questions that we can decompose using our Design Challenge rubric.
Trying to define "gene expression"
The first thing we need to do, however, is to define what it means when we say that a gene is "expressed". If the gene encodes a protein, one might reasonably propose that "expression" of a gene means how much functional protein the cell makes. But what if the gene does not encode a protein but some functional RNA. Then, in this case, "expressed might mean how much of the functional RNA the cell makes. Yet another person might reasonably suggest that "expression" just refers to the initial step in creating a copy of the genomic information. By that definition, one might want to count how many full-length transcripts are being made. Is it the number of end products encoded by the genomic information or is it the number of reads of the information important to describe "expression" properly. Unfortunately, in practice, we often find that the definition depends on the context of the discussion. Keep that in mind. For the sake of making sure that we are talking about the same thing, in Bis2A we'll try to use the term "expression" primarily to describe the creation of the final functional product(s). Depending on the specific case, the final product may be a protein or RNA species.
The design challenge of regulating gene expression
To drive this discussion from a design challenge perspective, we can formally stipulate that the "big problem" we are interested in understating is that of regulating protein abundance in a cell. Problem: The cell must regulate the abundance of each functional protein. We can then start by posing subproblems:
Let's take a moment, though, first to reload a few ideas. The process of gene expression requires multiple steps depending on what the fate of the final product will be. With structural and regulatory RNAs (i.e. tRNA, rRNA, snRNA, etc.) the process requires that a gene be transcribed and that any needed post-transcriptional processing takes place. With a protein-coding gene, the transcript must also be translated into protein and, if required, modifications to the protein must also be made. Both transcription and translation are multi-step processes, and most of those sub-steps are also potential sites of control.
Some subproblems might therefore be:
- It is reasonable to postulate that there must be some mechanism(s) to regulate the first step of this multi-step process, the initiation of transcription (just getting things started). So, we could state, "we need a mechanism to regulate the initiation of transcription." We could also turn this into a question and ask, "how can the initiation of transcription be accomplished"?
- We can use similar thinking to state, "we need a mechanism for regulating the end of transcription" or to ask "how is transcription terminated?"
- Using this convention we can state, "we need to regulate the initiation of translation and the stop of translation".
- We've talked only about the synthesis of protein and RNA. It is also reasonable to state, "we need a mechanism to regulate the degradation of RNA and protein."
Focusing on transcription
In this course we begin by focusing primarily on examining the first couple of problems/questions, regulating transcription initiation and termination - from genomic information to a functional RNA, either ready as is (e.g. with a functional RNA) or ready for translation. This allows us to examine some fundamental concepts regarding the regulation of gene expression and to examine a few real examples of those concepts in action.
Subproblems for transcription and the activity or RNA polymerase
Let us consider a protein-coding gene and work through some logic. We start by imagining a simple case, where a protein-coding gene is encoded by a single contiguous stretch of DNA. We know that to transcribe this gene an RNA polymerase will need to be recruited to the start of the coding region. The RNA polymerase is not "smart" per se. There needs to be some mechanism, based on chemical logic, to help recruit the RNA polymerase to the start of the protein-coding gene. Likewise, if this process is to be regulated, there needs to be some mechanism, or mechanisms to dictate when an RNA polymerase should be recruited to the start of a gene, when it should not, and/or if it is recruited to the DNA whether it should begin transcription and how many times this process should happen. Note, that the previous sentence, has several distinct subproblems/questions (e.g. when is the polymerase recruited?; if recruited, should it start transcription?; if it starts transcription, how many times should this process repeat?). We can also reasonably infer, that there will need to be some mechanisms to "instruct" (more anthropomorphisms) the polymerase to stop transcription. Finally, since the role of transcription is to create RNA copies of the genome segments, we should also consider problems/questions related to other factors that influence the abundance of RNA, like mechanisms of degradation. There must be some mechanisms, and these mechanisms will probably involve regulating this process.
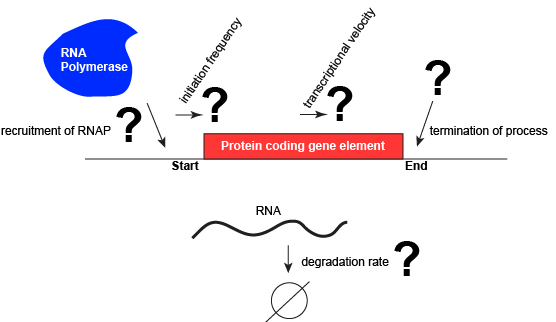
A schematic showing a protein-coding gene and some questions or problems that we need to ask ourselves or problems we need to know solutions for if we are to understand how regulation of the transcriptional portion of the gene's expression is regulated. Attribution: Marc T. Facciotti (own work)
Activation and Repression of Transcription
Some basics
Let us consider a protein-coding gene and work through some logic. We start by imagining a simple case, where a protein-coding gene is encoded by a single contiguous stretch of DNA. We know that to transcribe this gene an RNA polymerase will need to be recruited to the start of the coding region. The RNA polymerase is not "smart" per se. There needs to be some mechanism, based on chemical logic, to help recruit the RNA polymerase to the start of the protein-coding gene. Likewise, if this process is to be regulated, there needs to be some mechanism, or mechanisms to dictate when an RNA polymerase should be recruited to the start of a gene, when it should not, and/or if it is recruited to the DNA, whether it should begin transcription and how many times this process should happen. Note, that the previous sentence, has several distinct subproblems/questions (e.g. when is the polymerase recruited?, if recruited should it start transcription?, if it starts transcription, how many times should this process repeat?). We can also reasonably infer, that there will need to be some mechanisms to "instruct" (more anthropomorphisms) the polymerase to stop transcription.
Recruiting RNA polymerase to specific sites
To initiate transcription, the RNA polymerase must be recruited to a segment of DNA near the start of a region of DNA encoding a functional transcript. The function of the RNA polymerase as described so far, however, is not to bind specific sequences but to move along any segment of DNA. Recruiting the polymerase to a specific site therefore seems contradictory to its usual behavior. Explaining this contradiction requires us to invoke something new. Either transcription can start anywhere and just those events that lead to a full productive transcript do anything useful or something other than the RNA polymerase itself helps to recruit the enzyme to the beginning of a gene. The latter, we now take for granted, is the case.
The recruitment of the RNA polymerase is mediated by proteins called general transcription factors. In bacteria, they have a special name: sigma factors. In archaea, they are called TATA binding protein and transcription factor IIB. In eukaryotes, relatives of the archaeal proteins function with many others to recruit the RNA polymerase. The general transcription factors have at least two basic functions: (1) They can chemically recognize a specific sequence of DNA and (2) they can bind the RNA polymerase. Together these two functions of general transcription factors solve the problem of recruiting an enzyme that is otherwise not capable of binding a specific DNA site. In some texts, the general transcription factors (and particularly the sigma factor varieties) are said to be part of the RNA polymerase. While they are part of the complex when they help to target the RNA polymerase they do not continue with the RNA polymerase after it starts transcription.
The DNA site that recruits an RNA polymerase has a special name. We call it a promoter. While the DNA sequences of different promoters need not be exactly the same, different promoter sequences typically have some special chemical properties in common. Obviously, one property is that they can associate with an RNA polymerase. In addition, the promoter usually has a DNA sequence that facilitates the dissociation of the double stranded DNA such that the polymerase can begin reading and transcription the coding region. (Note: technically we could have broken down the properties of the promoter into design challenge subproblems. Here we skipped it, but you should still be able to step backwards and create the problem statements and or relevant questions once you find out about promoters).
In nearly all cases, but particularly in eukaryotic systems the complex of proteins that assembles with the RNA polymerase at promoters (typically called the pre-initiation complex) can number in the tens of proteins. Each of these other proteins has a specific function, but this is far too much detail to dive into for Bis2A.
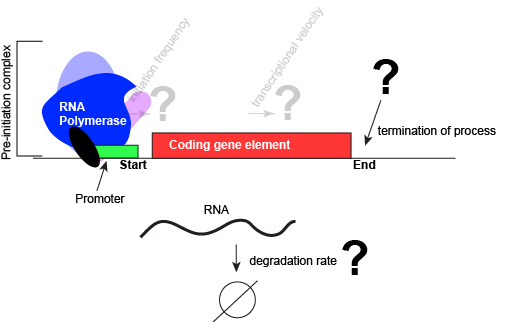
An abstract model of a generic transcriptional unit shown above with the addition of a promoter and PIC. Questions noted earlier that will probably not be covered in Bis2a have been grayed out. Attribution: Marc T. Facciotti (own work)
States of a regulated promoter
Since promoters recruit an RNA polymerase these sites and the assembly of the pre-initiation complex are obvious sites for regulating the first steps of gene expression. At the level of transcription initiation, we often classify promoter into one of three classes. We call the first constitutive. Constitutive promoters are generally not regulated very strongly. Their base state is "on". When the constitutive transcription from a promoter is very high (relative to most other promoters), we will colloquially call that promoter a "strong constitutive" promoter. If the amount of transcription from a constitutive promoter is low (relative to most other promoters) we call that promoter a "weak constitutive" promoter.
A second way to classify promoters by the use of the term activated or equivalently induced. These interchangeable terms are used to describe promoters sensitive to some external stimulus and respond to said stimulus by increasing transcription. Activated promoters have a base state exhibits little to no transcription. Transcription is then "activated" in response to a stimulus - the stimulus turns the promoter "on".
Finally, the third term used to classify promoters is by the use of the term repressed. These promoters also respond to stimuli but do so by decreasing transcription. The base state for these promoters is some basal level of transcription, and the stimulus acts to turn down or repress transcription. Transcription is "repressed" in response to a stimulus - the stimulus turns the promoter "off".
The examples given above assumed that a single stimulus acts to regulate promoters. While this is the simplest case, many promoters may integrate different information and may be alternately activated by some stimuli and repressed by other stimuli.
Possible NB Discussion  Point
Point
In Bis2A, you have learned about how important it is for the cell to be able to regulate its biology, with several examples of regulatory mechanisms. Given this, can you explain why it is advantageous to still have constitutive promoters that are generally not regulated very strongly? What kinds of genes would you expect to have a strong constitutive promoter? What about a weak constitutive promoter?
Transcription factors help to regulate the behavior of a promoter
How are promoters sensitive to external stimuli? Mechanistically, in both activation and repression require regulatory proteins to change the transcriptional output of the gene being observed. The proteins responsible for helping to regulate expression are generally called transcription factors. The specific DNA sequences bound by transcription factors are often called operators and most times the operators are very close to the promoter sequences.
Here's where the nomenclature gets potentially confusing - particularly when comparing across bacterial and eukaryotic research. In bacterial research, if the transcription factor acts by binding DNA and the RNA polymerase in a way that increases transcription, then it is typically called an activator. If, by contrast, the transcription factor acts by binding DNA to repress or decrease transcription of the gene then it is called a repressor.
Why are the classifications of activator and repressor potentially problematic? These terms describe the proteins with idealized single functions. While this may be true with some transcription factors, in reality other transcription factors may act to activate gene expression in some conditions while repressing in other conditions. Some transcription factors will act to modulate expression either up or down depending on context rather than shutting transcription "off" or turning it completely "on". To circumvent some of this confusion, some of your instructors prefer to avoid using the terms activator and repressor and instead prefer to discuss the activity of transcription various transcription factors as either a positive or a negative influence on gene expression in specific cases. If we use these terms, you might hear your instructor saying that the transcription factor in question ACTS LIKE/AS a repressor or that it ACTS LIKE/AS an activator, taking care not to call it simply an activator or repressor. It is more likely, however, that you will hear them say that a transcription factor is acting to positively or negatively influence transcription.
CAUTION: Depending on your instructor, you may cover a few real biological examples of positive and negative regulatory mechanisms. These specific examples will use the common names of the transcription factors - since the examples will typically be drawn from the bacterial literature, the names of the transcription factors may include the terms "repressor" or "activator". These names are relics of when they were first discovered. Try to spend more time examining the logic of how the system works than trying to commit to memory any special properties of that specific protein to all other cases with the same label. Just because we label a protein as a repressor does not mean that it only acts as a negative regulator in all cases or that it interacts with external signals in the same was as the example.
Possible NB Discussion  Point
Point
What types of interactions do you think happen between the amino acids of the transcription factor and the double helix of the DNA? How do transcription factors recognize their binding site on the DNA?
Allosteric Modulators of Regulatory Proteins
The activity of many proteins, including regulatory proteins and various transcription factors, can be allosterically modulated various factors, including by the relative abundance of small molecules in the cell. We often refer these small molecules as inducers or co-repressorsor co-activators and are often metabolites, such as lactose or tryptophan or small regulatory molecules, such as cAMP or GTP. These interactions allow the TF to respond to environmental conditions and to modulate its function accordingly. It helps to decide whether to transcribe a gene depending on the abundance of the environmental signal.
Let us imagine a negative transcriptional regulator. In the most simple case we've considered so far, transcription of a gene with a binding site for this transcription factor would be low when the TF is present and high when the TF is absent. We can now add a small molecule to this model. Here the small molecule can bind the negative transcriptional regulator through sets of complementary hydrogen and ionic bonds. In this first example we will consider the case where the binding of the small molecule to the TF induces a conformational change to the TF that severely reduces its ability to bind DNA. If this is the case, the negative regulator - once bound by its small molecule - would release from the DNA. This would thereby relieve the negative influence and lead to increased transcription. This regulatory logic might be appropriate to have evolved in the following scenario: a small molecule food-stuff is typically absent from the environment. Therefore, genes encoding enzymes that will degrade/use that food should be kept "off" most of the time to preserve the cellular energy that their synthesis would use. A negative regulator could accomplish this. When the food-stuff appears in the environment, it would be appropriate for the enzymes responsible for its processing to be expressed. The food-stuff could then act by binding to the negative regulator, changing the TF's conformation, causing its release from the DNA and turning on transcription of the processing enzymes.
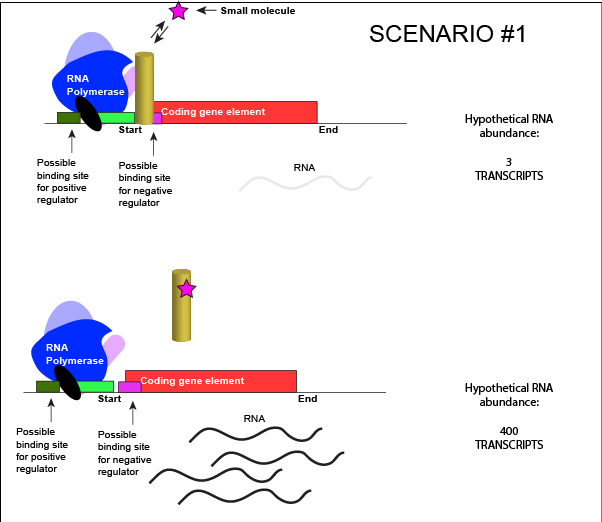
An abstract model of a generic transcriptional unit regulated by a negative regulator whose activity is modulated by a small molecule (depicted by a star). Here, the binding of the small molecule causes the TF to release from the DNA. Attribution: Marc T. Facciotti (own work)
We can consider a second model for how a negatively acting TF might interact with a small molecule. Here, the TF alone cannot bind its regulatory site to the DNA. However, when a small molecule binds to the TF a conformational change occurs that reorients DNA binding amino-acids into the "correct" orientation for DNA binding. The TF-small molecule complex now binds to the DNA and acts to influence transcription negatively.
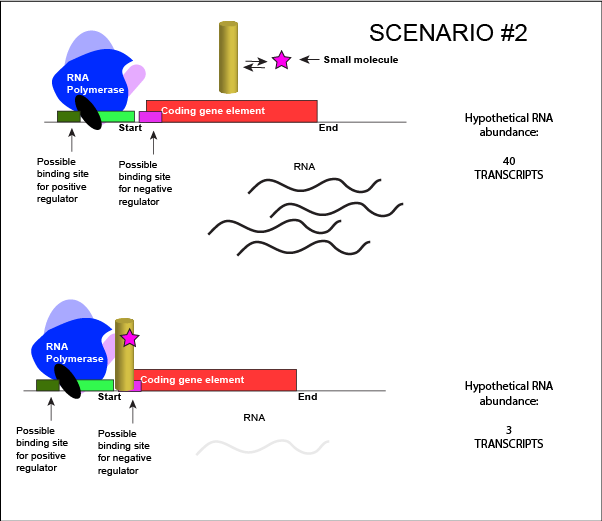
An abstract model of a generic transcriptional unit regulated by a negative regulator whose activity is modulated by a small molecule (depicted by a star). Here, binding of the small molecule causes the TF to bind to the DNA. Attribution: Marc T. Facciotti (own work)
Note how the activity of the TF can be modulated in distinctly different ways by a small molecule. Depending on the protein, the binding of this external signal can either cause binding of the TF-small molecule complex to DNA OR binding of the small molecule can cause the release of the TF-small molecule complex from the DNA. We can work up the same kinds of examples for a positive regulator.
In both cases proposed above, the binding of a small molecule to a TF will depend on how strongly the TF interacts with the small molecule. This will depend on the types and spatial orientation of the protein's chemical functional groups, their protonation states (if applicable), and the complementary functional groups on the small molecule. It should not be surprising, therefore, to learn that the binding of the small molecule to the TF will depend on various factors, including but not limited to the relative concentrations of small-molecule and TF and pH.
Is it positive or negative regulation?
Resolving a common point of confusion
It is not uncommon for many Bis2a students to be slightly confused about how to determine if a transcription factor is acting as a positive or negative regulator. This confusion often comes after a discussion of the modes that the stimulus (i.e. small molecule) can influence the activity of a transcription factor. This is not too surprising. In the examples above, the binding of an effector molecule to a transcription factor could have one of two different effects: (1) binding of the effector molecule could induce a DNA-bound transcription factor to release from its binding site, derepressing a promoter, and "turning on" gene expression. (2) binding of the effector molecule to the transcription factor could induce the TF to bind to its DNA binding site, repressing transcription, and "turning off" gene expression. In the first case, it might appear that the TF is acting to regulate expression positively, while in the second example it might appear that the TF acts negatively.
However, in both examples above, the TF is acting as a negative regulator. To determine this, we look at what happens when the TF binds DNA (whether a small molecule is bound to the TF or not). In both cases, binding of the TF to DNA represses transcription. The TF is therefore acting as a negative regulator. We can conduct a similar analysis for positively acting TFs.
Note that sometimes a TF may act as a positive regulator at one promoter and negative regulator at a different promoter so describing the behavior of the TF on a per case basis is often important (reading too much from the name it has been assigned can mislead sometimes). Other TF proteins can act alternately as both positive or negative regulators of the same promoter depending on conditions. Again, describing the behavior of the TF specifically for each case is advised.
A genetic test for positive or negative regulatory function of a TF
How does one determine if a regulatory protein functions in a positive or negative way? A simple genetic test is to ask "what happens to expression if the regulatory protein is absent?" This can be accomplished by removing the coding gene for the transcription factor from the genome. If a transcription factor acts positively, then its presence is required to activate transcription. In its absence, there is no regulatory protein, therefore no activation, and the outcome is lower transcription levels of a target gene. The opposite is true for a transcription factor acting negatively. In its absence expression should be increased, because the gene keeping expression low is no longer around.
Summary of gene regulation
In the preceding text we have examined several ways to solve some design challenges associated with regulating the amount of transcript that is created for a single coding region of the genome. We have looked in abstract terms at some processes responsible for controlling the initiation of transcription, how these may be made sensitive to environmental factors, and very briefly at the processes that terminate transcription and handle the active degradation of RNA. We have also discussed how Nature can quantitatively tune each of these processes to be "stronger" or "weaker” and that this tuning of "strength" (e.g. promoter strength, degradation rates, etc.) Influence the behavior of the overall process in potentially functionally important ways.
Examples of Bacterial Gene Regulation
This section describes two examples of transcriptional regulation in bacteria. These are presented as illustrative examples. Use these examples to learn some basic principles about mechanisms of transcriptional regulation. Be on the lookout in class, in discussion, and in the study-guides for extensions of these ideas and use these to explain the regulatory mechanisms used for regulating other genes.
Gene Regulation Examples in E. coli
The DNA of bacteria and archaea are usually organized into one or more circular chromosomes in the cytoplasm. The dense aggregate of DNA that can be seen in electron micrographs is called the nucleoid. In bacteria and archaea, genes, whose expression needs to be tightly coordinated (e.g. genes encoding proteins involved in the same biochemical pathway) are often grouped closely together in the genome. When the expression of multiple genes is controlled by the same promoter and a single transcript is produced these expression units are called operons. For example, in the bacterium Escherichia coli all the genes needed to utilize lactose are encoded next to one another in the genome. We call this arrangement the lactose (or lac) operon. In many bacteria and archaea nearly 50% of all genes are encoded into operons of two or more genes.
The Role of the Promoter
The first level of control of gene expression is at the promoter itself. Some promoters recruit RNA polymerase and turn those DNA-protein binding events into transcripts more efficiently than other promoters. This intrinsic property of a promoter, it's ability to produce transcript at a particular rate, is referred to as promoter strength. The stronger the promoter, the more RNA is made in any given time period. Promoter strength can be "tuned" by Nature in very small or very large steps by changing the nucleotide sequence the promoter (e.g. mutating the promoter). This results in families of promoters with different strengths that can be used to control the maximum rate of gene expression for certain genes.
UC Davis Undergraduate Connection:
A group of UC Davis students interested in synthetic biology used this idea to create synthetic promoter libraries for engineering microbes as part of their design project for the 2011 iGEM competition.
Example #1: Trp Operon
E. coli, like all organisms, needs to either synthesize or consume amino acids to survive. The amino acid tryptophan is one such amino acid. E. coli can either import tryptophan from the environment (eating what it can scavenge from the world around it) or synthesize tryptophan de novo using enzymes encoded by five genes. These five genes reside next to each other in the E. coligenome in what we call the tryptophan (trp) operon (Figure below). Just before the coding region is the transcriptional start site. This is, as the name implies, the location where the RNA polymerase starts a new transcript. The promoter sequence is further upstream of the transcriptional start site. A DNA sequence called an "operator" is also encoded between the promoter and the first trp coding gene. This operator is the DNA sequence to which the transcription factor protein will bind.
If tryptophan is present in the environment, then E. coli need not synthesize it and the switch controlling the activation of the genes in the trp operon switches off. However, when environmental tryptophan availability is low, the switch controlling the operon is turned on, transcription is initiated, the genes are expressed, and the organism synthesizes tryptophan.
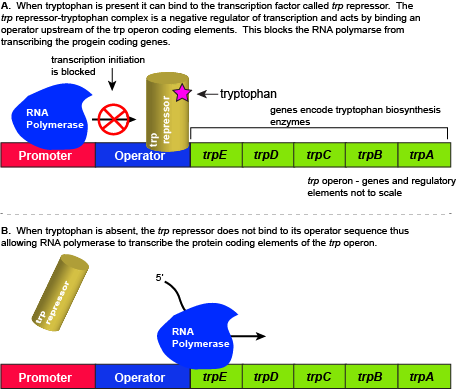
The five genes that required to synthesize tryptophan in E. coli group next to each other in the trp operon. When tryptophan is plentiful, two tryptophan molecules bind to the transcription factor and allow the TF-tryptophan complex to bind at the operator sequence. This physically blocks the RNA polymerase from transcribing the tryptophan biosynthesis genes. When tryptophan is absent, the transcription factor does not bind to the operator and the genes are transcribed. Attribution: Marc T. Facciotti (own work)
Possible NB Discussion Point
Suppose nature took a different approach to regulating the trp operon. Propose a method for regulating the expression of the trp operon with a positive regulator instead of a negative regulator. Describe how this might work. (Hint: we ask this kind of question on exams)
Example #2: The lac operon
In this example, we examine the regulation of genes encoding proteins whose physiological role is to import and assimilate the disaccharide lactose, the lac operon. The story of regulating lac operon is a common example used in many introductory biology classes to illustrate basic principles of inducible gene regulation.
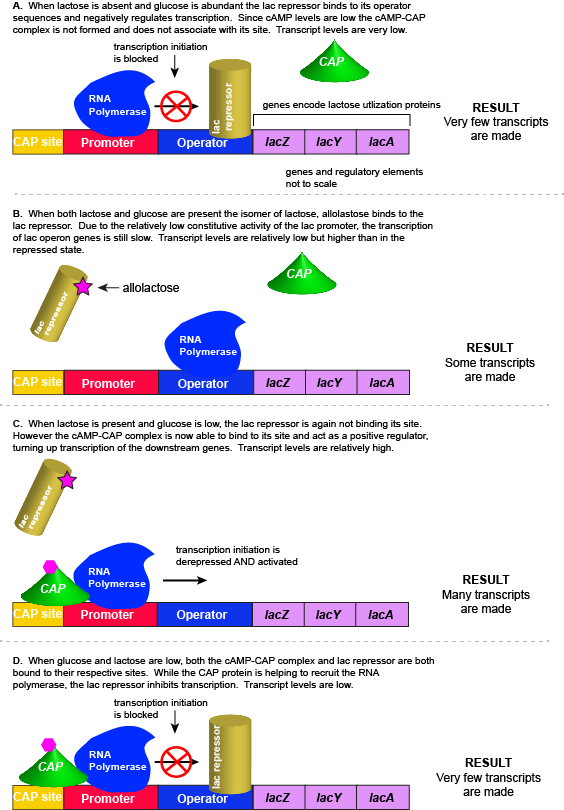
Lactose is a disaccharide composed of the hexoses glucose and galactose. We commonly encounter lactose in milk and some milk products. As one can imagine, the disaccharide can be an important food-stuff for microbes that can use its two hexoses. E. colican use multiple different sugars as energy and carbon sources, including lactose and the lac operon is a structure that encodes the genes necessary to acquire and process lactose from the local environment. E. coli, however, does not frequently encounter lactose, and therefore the genes of the lac operon must typically be repressed (i.e. "turned off") when lactose is absent. Driving transcription of these genes when lactose is absent would waste precious cellular energy. By contrast, when lactose is present, it would make logical sense for the genes responsible for using the sugar to be expressed (i.e. "turned on"). So far, the story is very similar to that of the tryptophan operon described above.
However, there is a catch. Experiments conducted in the 1950's by Jacob and Monod showed that E. coli prefers to use all the glucose present in the environment before it uses lactose. This means that the mechanism used to decide whether or not to express the lactose utilization genes must be able to integrate two types of information (1) the concentration of glucose and (2) the concentration of lactose. While this could theoretically be accomplished in multiple ways, we will examine how the lac operon accomplishes this by using multiple transcription factors.
The transcriptional regulators of the lac operon
1. The lac repressor - a direct sensor of lactose
As noted, the lac operon normally has very low to no transcriptional output in the absence of lactose. This is because of two factors: (1) the constitutive promoter strength for the operon is relatively low and (2) the constant presence of the LacI repressor protein negatively influences transcription. This protein binds to the operator site near the promoter and blocks RNA polymerase from transcribing the lac operon genes. By contrast, if lactose is present, lactose will bind to the LacI protein, inducing a conformational change that prevents LacI-lactose complex from binding to its binding sites. Therefore, when lactose is present, the negative regulatory LacI is not bound to its binding site and transcription of lactose using genes can proceed.
2. CAP protein - an indirect sensor of glucose
In E. coli, when glucose levels drop, the small molecule cyclic AMP (cAMP) accumulates in the cell. cAMP is a common signaling molecule that is involved in glucose and energy metabolism in many organisms. When glucose levels decline in the cell, the increasing concentrations of cAMP allow this compound to bind to the positive transcriptional regulator called catabolite activator protein (CAP) - also referred to as CRP. cAMP-CAP complex has many sites throughout the E. coli genome and many of these sites are located near the promoters of many operons that control the processing of various sugars.
In the lac operon, the cAMP-CAP binding site is located upstream of the promoter. Binding of cAMP-CAP to the DNA helps to recruit and keep RNA polymerase to the promoter. The increased occupancy of RNA polymerase to its promoter, in turn, results in increased transcriptional output. Here, the CAP protein is acting as a positive regulator.
Putting it all together: Inducing expression of the lac operon
For the lac operon to be activated, two conditions must be met. First, the level of glucose must be very low or non-existent. Second, lactose must be present. Only when glucose is absent and lactose is present, will the lac operon be transcribed. When this condition is achieved the LacI-lactose complex dissociates the negative regulator from near the promoter, freeing the RNA polymerase to transcribe the operon's genes. High cAMP (indirectly indicative of low glucose) levels trigger the formation of the CAP-cAMP complex. This TF-inducer pair now bind near the promoter and act to positively recruit the RNA polymerase. This added positive influence boosts transcriptional output and lactose can be efficiently utilized. The mechanistic output of other combinations of binary glucose and lactose conditions are described in the table below and in the figure that follows.
| Signals that Induce or Repress Transcription of the lac Operon | ||||
|---|---|---|---|---|
| Glucose | CAP binds | Lactose | Repressor binds | Transcription |
| + | - | - | + | No |
| + | - | + | - | Some |
| - | + | - | + | No |
| - | + | + | - | Yes |
Link to external resources
Gene Regulation and the Order of the Operon
https://www.youtube.com/watch?v=h_1QLdtF8d0
Regulation of Gene Expression: Operons, Epigenetics, and Transcription Factors
https://www.youtube.com/watch?v=J9jhg90A7Lw
Prokaryotic Gene Regulation: Lac Operon
https://www.youtube.com/watch?v=-4Jr84qyKPo