1.1: The Structure of DNA
- Page ID
- 25720
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Learning Objectives
- Identify the sugar, phosphate, nitrogenous base, 5' and 3' carbons in a nucleotide and the key difference between DNA and RNA.
- Explain the structure of the double helix, including the role of hydrogen bonds and covalent (phosphodiester) bonds.
- Explain why the abundance of A is roughly equal to T and G is roughly equal to C in DNA.
- Using the rules of base pairing, predict the complementary (or anti-parallel) strand of DNA for a given sequence.
Nucleic acids are the most important macromolecules for the continuity of life. They carry the genetic blueprint of a cell and carry instructions for the functioning of the cell.
DNA and RNA
The two main types of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). DNA is the genetic material found in all living organisms, ranging from single-celled bacteria to multicellular mammals. DNA is found in the nucleus of eukaryotes and in the organelles, chloroplasts, and mitochondria. In prokaryotes, the DNA is not enclosed in a membranous envelope.
The entire genetic content of a cell is known as its genome, and the study of genomes is genomics. In eukaryotic cells but not in prokaryotes, DNA forms a complex with histone proteins to form chromatin, the substance of eukaryotic chromosomes. A chromosome may contain tens of thousands of genes. Many genes contain the information to make protein products; other genes code for RNA products. DNA controls all of the cellular activities by turning the genes “on” or “off.”
The other type of nucleic acid, RNA, is mostly involved in protein synthesis. The DNA molecules never leave the nucleus but instead use an intermediary to communicate with the rest of the cell. This intermediary is the messenger RNA (mRNA). Other types of RNA—like rRNA, tRNA, and microRNA—are involved in protein synthesis and its regulation.
DNA and RNA are made up of monomers known as nucleotides. The nucleotides combine with each other to form a polynucleotide, DNA or RNA. Each nucleotide is made up of three components: a nitrogenous base, a pentose (five-carbon) sugar, and a phosphate group (Figure \(\PageIndex{1}\)). Each nitrogenous base in a nucleotide is attached to a sugar molecule, which is attached to one or more phosphate groups.
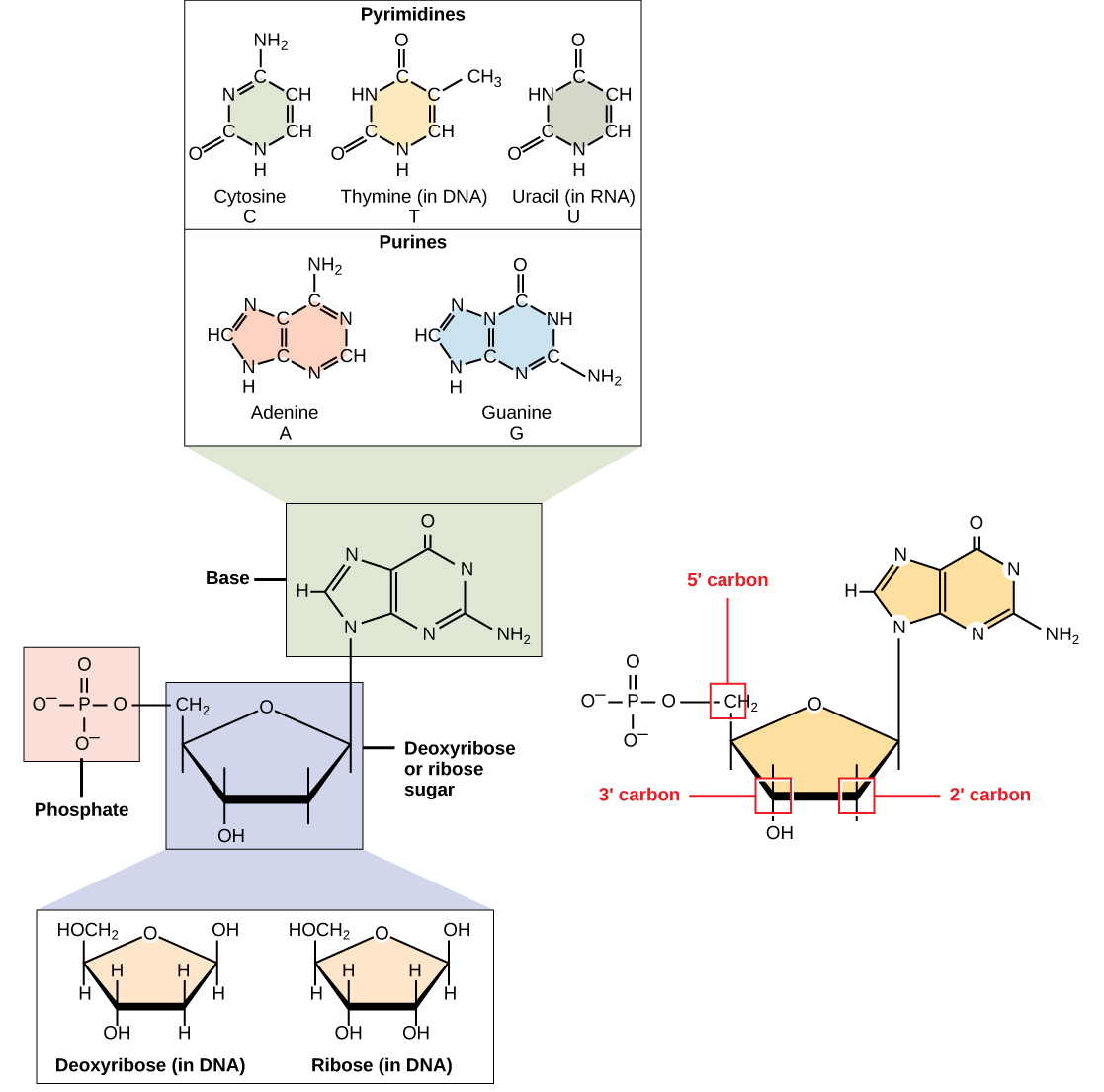
The nitrogenous bases, important components of nucleotides, are organic molecules and are so named because they contain carbon and nitrogen. They are bases because they contain an amino group that has the potential of binding an extra hydrogen, and thus, decreasing the hydrogen ion concentration in the environment, making it more basic. Each nucleotide in DNA contains one of four possible nitrogenous bases: adenine (A), guanine (G) cytosine (C), and thymine (T).
Adenine and guanine are classified as purines. The primary structure of a purine is two carbon-nitrogen rings. Cytosine, thymine, and uracil are classified as pyrimidines which have a single carbon-nitrogen ring as their primary structure (Figure \(\PageIndex{1}\)). Each of these basic carbon-nitrogen rings has different functional groups attached to it. In molecular biology shorthand, the nitrogenous bases are simply known by their symbols A, T, G, C, and U. DNA contains A, T, G, and C whereas RNA contains A, U, G, and C.
The pentose sugar in DNA is deoxyribose, and in RNA, the sugar is ribose (Figure \(\PageIndex{1}\)). The difference between the sugars is the presence of the hydroxyl group on the second carbon of the ribose and hydrogen on the second carbon of the deoxyribose (so deoxyribose is "missing" an -OH group). The carbon atoms of the sugar molecule are numbered as 1′, 2′, 3′, 4′, and 5′ (1′ is read as “one prime”).
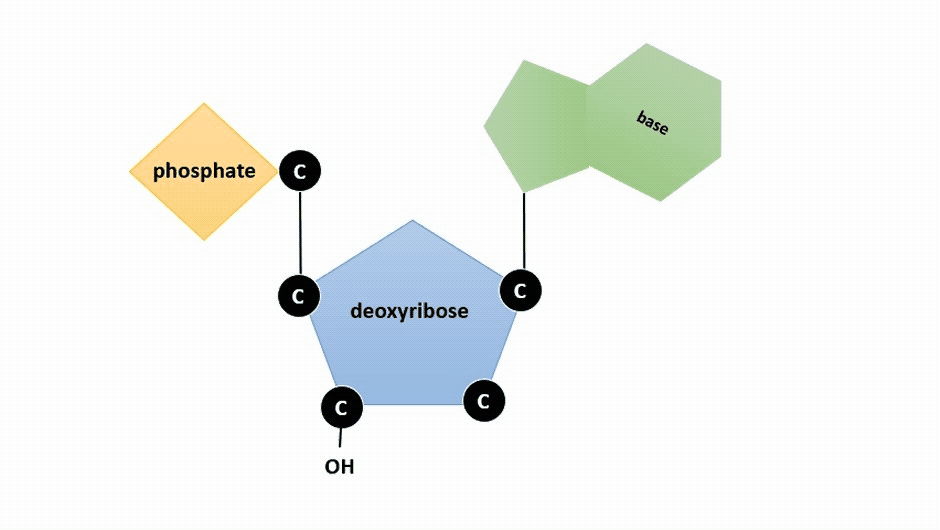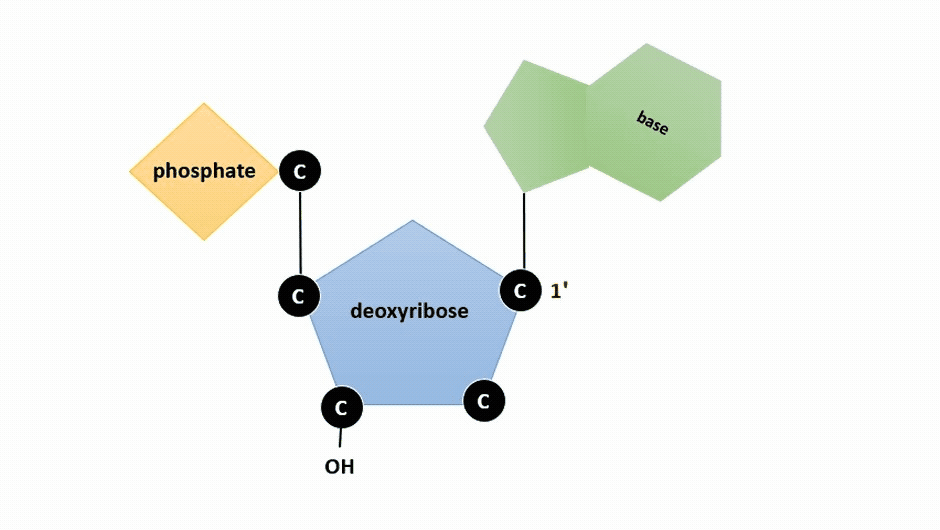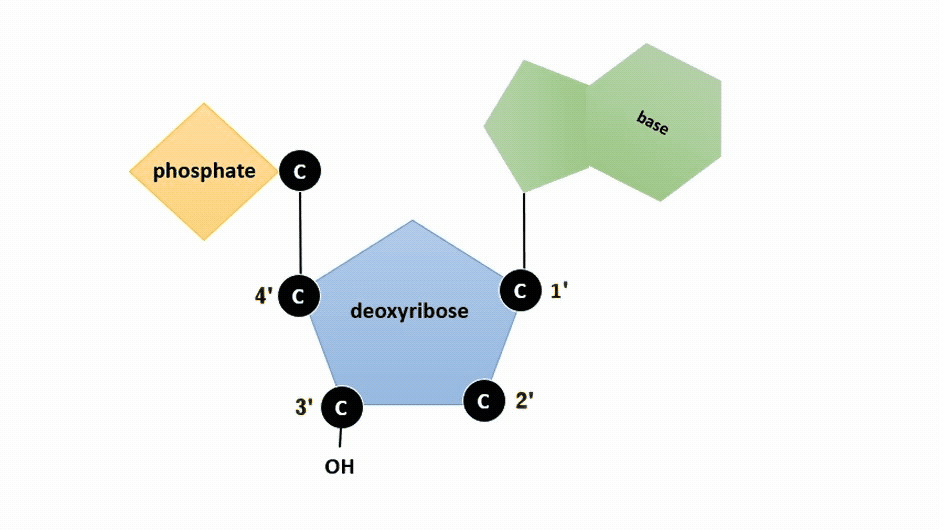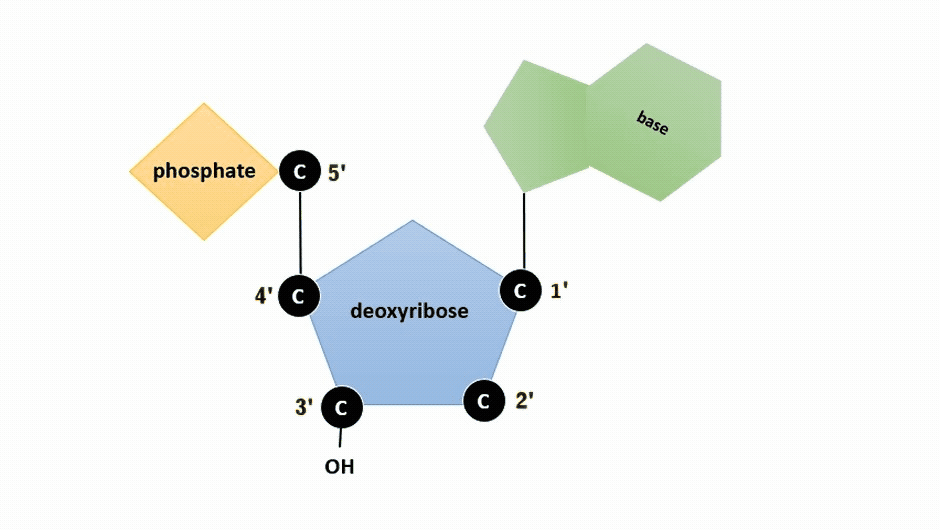
Figure \(\PageIndex{2}\): Animation highlighting carbons 1' through 5' on dexoyribose. (SWLeacock)
The phosphate residue is attached to the hydroxyl group of the 5′ carbon of one sugar and the hydroxyl group of the 3′ carbon of the sugar of the next nucleotide, which forms a 5′–3′ phosphodiester linkage. The phosphodiester linkage is not formed by simple dehydration reaction like the other linkages connecting monomers in macromolecules: its formation involves the removal of two phosphate groups. A polynucleotide may have thousands of such phosphodiester linkages.
Chargaff’s Rules
When Watson and Crick set out in the 1940’s to determine the structure of DNA, it was already known that DNA is made up of a series four different types of molecules, called bases or nucleotides: adenine (A), cytosine (C), thymine (T), guanine (G). Watson and Crick also knew of Chargaff’s Rules, which were a set of observations about the relative amount of each nucleotide that was present in almost any extract of DNA. Chargaff had observed that for any given species, the abundance of A was the same as T, and G was the same as C. This was essential to Watson & Crick’s model.
Example \(\PageIndex{1}\)
Chargaff determined the composition of nucleic acids in samples from a variety of species, including prokaryotes and eukaryotes. In one bacterial sample, the proportion of adenine was 15.5% (data adapted from Vischer et al, 1949). What proportion of guanine would have been present in this sample and why?
Solution
Because A pairs with T, the amount of T should be roughly equal to A, or approximately 15.5% percent. Thus, A + T = 15.5 + 15.5 = 31%.
The percent of G + C = 100% - 31% = 69%. Because G pairs with C, the amount of each of these should be roughly equal, so approximately 34.5% each.
Query \(\PageIndex{1}\)
DNA Double-Helix Structure
Using proportional metal models of the individual nucleotides, Watson and Crick deduced a structure for DNA that was consistent with Chargaff’s Rules and with x-ray crystallography data that was obtained (with some controversy) from another researcher named Rosalind Franklin. In Watson and Crick’s famous double helix, each of the two strands contains DNA bases connected through covalent (phosphodiester) bonds to a sugar-phosphate backbone. Because one side of each sugar molecule is always connected to the opposite side of the next sugar molecule, each strand of DNA has polarity: these are called the 5’ (5-prime) end and the 3’ (3-prime) end, in accordance with the nomenclature of the carbons in the sugars. The two strands of the double helix run in anti-parallel (i.e. opposite) directions, with the 5’ end of one strand adjacent to the 3’ end of the other strand. The double helix has a right-handed twist, (rather than the left-handed twist that is often represented incorrectly in popular media). The DNA bases extend from the backbone towards the center of the helix, with a pair of bases from each strand forming hydrogen bonds that help to hold the two strands together. Under most conditions, the two strands are slightly offset, which creates a major groove on one face of the double helix, and a minor groove on the other.
Rules of base pairing
- A pairs with T (or U if RNA)
- G pairs with C
Because of the structure of the bases, A can only form hydrogen bonds with T, and G can only form hydrogen bonds with C (remember Chargaff’s Rules). Each strand is therefore said to be complementary to the other, and so each strand also contains enough information to act as a template for the synthesis of the other. This complementary redundancy is important in DNA replication and repair. If the sequence of one strand is AATTGGCC, the complementary strand would have the sequence TTAACCGG. During DNA replication, each strand is copied, resulting in a daughter DNA double helix containing one parental DNA strand and a newly synthesized strand.
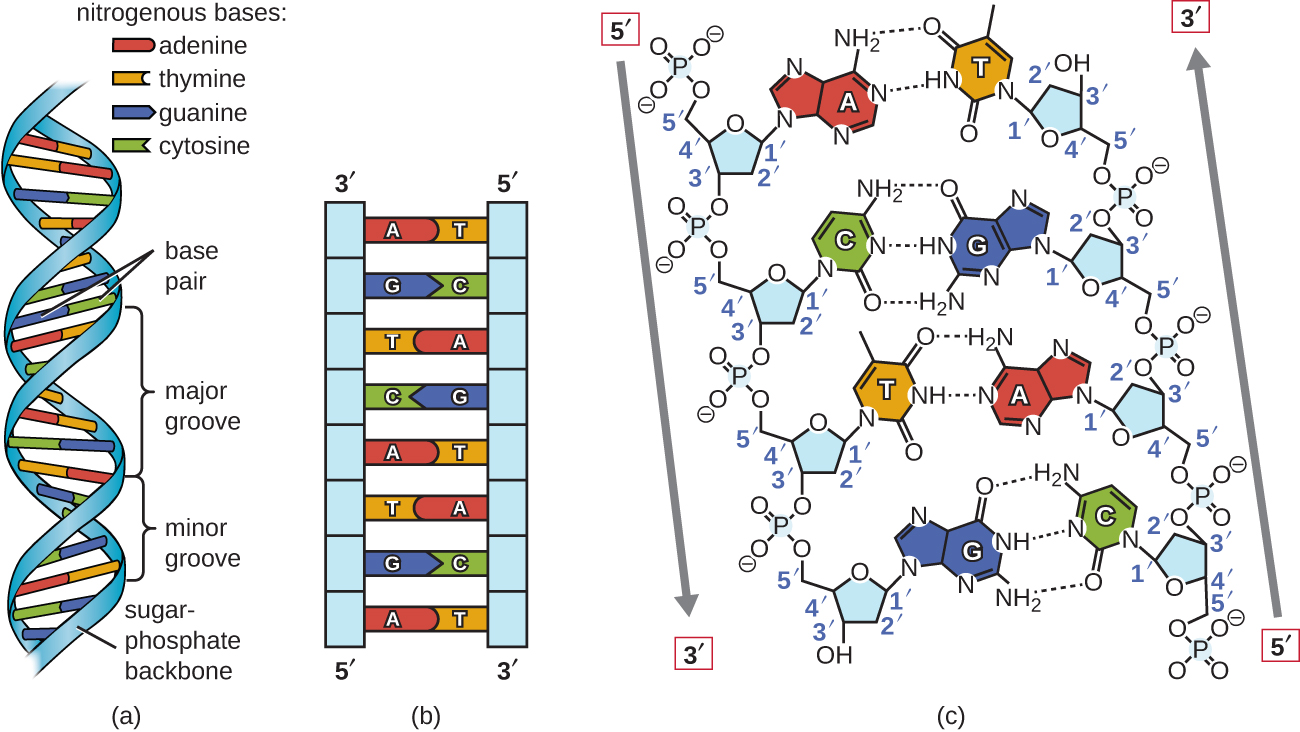
3D structure of a DNA double helix
Spin the double helix to see the orientation of the sugars and phosphates in the backbone (ribbon in the model), the base pairs, major and minor grooves! (PDB ID = 1bna https://www.rcsb.org/3d-view/1BNA)
Implications of DNA structure
As for most biological molecules, the structure is important to the function, and the function of DNA is to contain information. Important properties that are derived from the DNA structure are:
- A complementary strand can always be synthesized from a single strand, due to the arrangement of hydrogen bonds between GC and AT bases.
- Hydrogen bonds stabilize the double helix, but can be broken when DNA needs to be accessed.
- The order of bases contains the information needed to code for amino acids in proteins during translation.
- Even sequences of DNA that do not encode amino acids can still provide information by interacting with proteins that function in DNA packaging and regulation. The major and minor grooves of DNA may determine which sequences are visible to DNA interacting proteins.
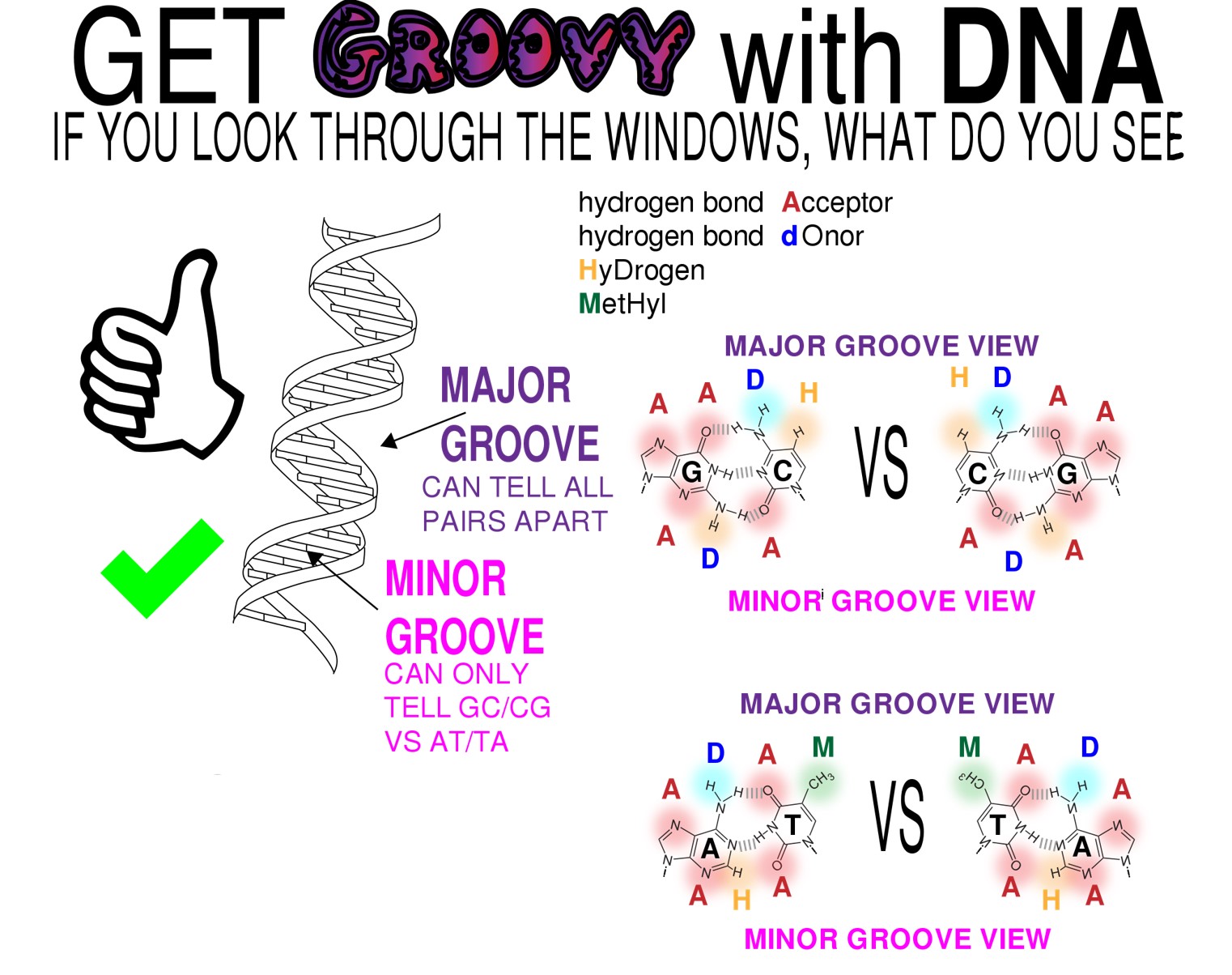
Figure \(\PageIndex{3}\): The significance of major and minor grooves in a DNA double helix. A DNA double helix twists in a right-handed fashion, just as the fingers on the right hand are "pointing" to the right when the right hand forms a "thumbs up." The predominant structure of a double helix results in major and minor grooves. The bases within the double helix interact with each other via hydrogen bonds, but the different bases pairs have different combinations atoms exposed in the major grooves. Proteins that recognize DNA sequences often do so by interacting with particular combinations of base pairs in major grooves based on these exposed atoms. In the minor grooves AT and TA base pairs appear the same , likewise GC and CG look the same in the minor groove. (Copyright By Biochemlife - Own work, CC BY-SA 4.0, https://commons.wikimedia.org/w/inde...curid=73107644)
Thinking ahead exercise \(\PageIndex{1}\)
A mutation occurs, and cytosine is replaced with adenine. What impact do you think this will have on the DNA structure?
- Answer
-
Adenine is larger than cytosine and will not be able to base pair properly with the guanine on the opposing strand, causing the double helix to bulge at that position. DNA repair enzymes may recognize the bulge and replace the incorrect nucleotide.
Thinking ahead exercise \(\PageIndex{2}\)
If the two DNA strands were connected by covalent bonds rather than hydrogen bonds, what problems might occur, if any?
- Answer
-
Covalent bonds are much stronger than hydrogen bonds and not as easily broken. If the DNA strands were connected by covalent bonds, then it would be much more difficult to unwind the double helix. This would be problematic for processes that require unwinding of the DNA molecule, such as replication and transcription.
Contributors and Attributions
Connie Rye (East Mississippi Community College), Robert Wise (University of Wisconsin, Oshkosh), Vladimir Jurukovski (Suffolk County Community College), Jean DeSaix (University of North Carolina at Chapel Hill), Jung Choi (Georgia Institute of Technology), Yael Avissar (Rhode Island College) among other contributing authors. Original content by OpenStax (CC BY 4.0; Download for free at http://cnx.org/contents/185cbf87-c72...f21b5eabd@9.87).
Access for free at https://openstax.org/books/biology/pages/1-introductionDr. Todd Nickle and Isabelle Barrette-Ng (Mount Royal University) The content on this page is licensed under CC SA 3.0 licensing guidelines.
References
VISCHER E, ZAMENHOF S, CHARGAFF E. Microbial nucleic acids; the desoxypentose nucleic acids of avian tubercle bacilli and yeast. J Biol Chem. 1949;177(1):429-438.


