1.1: Genetics Review
- Page ID
- 27164
Some Basic Terms
DNA is a double stranded nucleic acid polymer that is the primary hereditary material in most living organisms. Each strand of DNA is made up of covalently bound nucleotides. Each nucleotide is made up of a sugar, a phosphate, and a nitrogenous base. The covalent bond between nucleotides occurs between the sugar of one nucleotide and the phosphate of the next. In DNA, there is a single type of sugar but 4 types of bases. These bases - adenine, thymine, cytosine, and guanine - give each nucleotide its name (abbreviated A, T, C, and G).
The two strands of DNA are bound together by hydrogen bonds between nitrogenous bases. Each nitrogenous base has a "complementary" base. A pairs with T, and C pairs with G. If we think of a DNA strand having an orientation of sugar-phosphate-sugar-phosphate-sugar-phosphate then the second strand of the molecule has an orientation of phosphate-sugar-phosphate-sugar-phosphate-sugar. We call this type of orientation anti-parallel and we refer to the two strands as reverse complements. Reverse complementarity refers both to their opposite orientation (reverse) and the A=T G=C bonding pattern (complement).
RNA has a similar structure, though it is typically found as a single strand. It is capable of forming double strands (or even more complex structures, for example tRNA) through hydrogen bonding with reverse complement sequences. It can hydrogen bond with itself or with other RNA molecules.
The Central Dogma of Molecular Biology
For the most part when we think about Molecular Genetics we are thinking about making functional proteins from a gene encoded in DNA, though some RNA is functional on its own.
The production of protein from a DNA code happens via two highly-regulated processes: Transcription uses RNA polymerase to make a single stranded mRNA molecule from one strand of a double stranded DNA molecule. Translation uses a ribosome to make a peptide (part or all of a protein) from the mRNA (Figure 1).
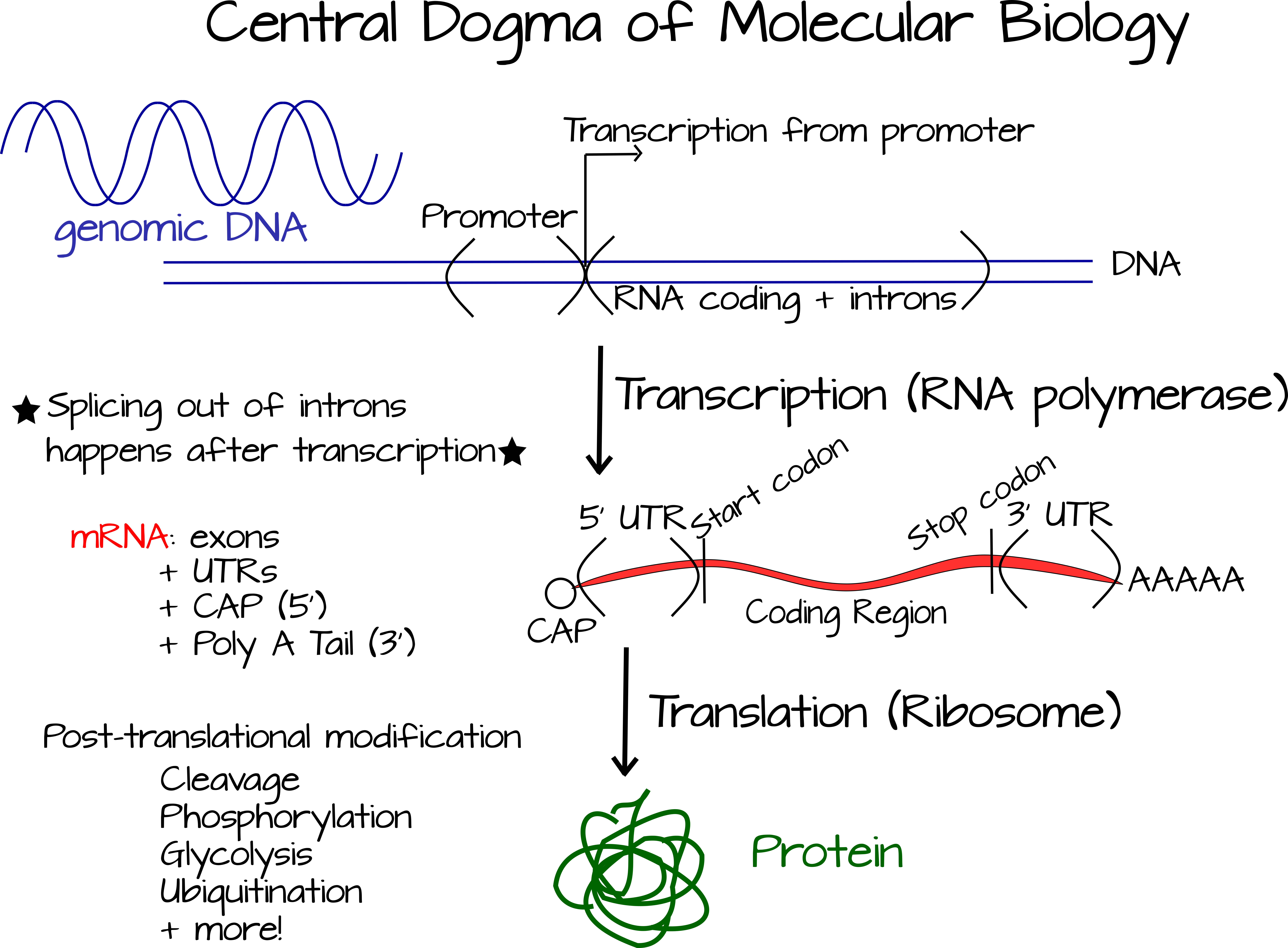
Quick side note because many people are confused by some of the terms that we use in genetics, evolution, and development. A gene refers to a piece of DNA that codes for RNA. This RNA is often an mRNA but can also be a tRNA, rRNA, snRNA, miRNA, or a different "non-coding" type of RNA. mRNA is RNA that codes for protein. Most of the time "gene expression" refers to the production of mRNAs and/or the proteins they code for. Gene expression is the process of using DNA to make a functional product. As you probably know, most of the cells in your body are genetically identical (more or less); they "share a genome." However, your cells have very different functions. These different functions are driven by different environments and different gene expression profiles - that is, the cells express different genes. For example, we might expect to find a lot of Myosin protein in muscle cells but a lot of Aquaporin protein in kidney cells.
The regulation of gene expression (epigenetics)
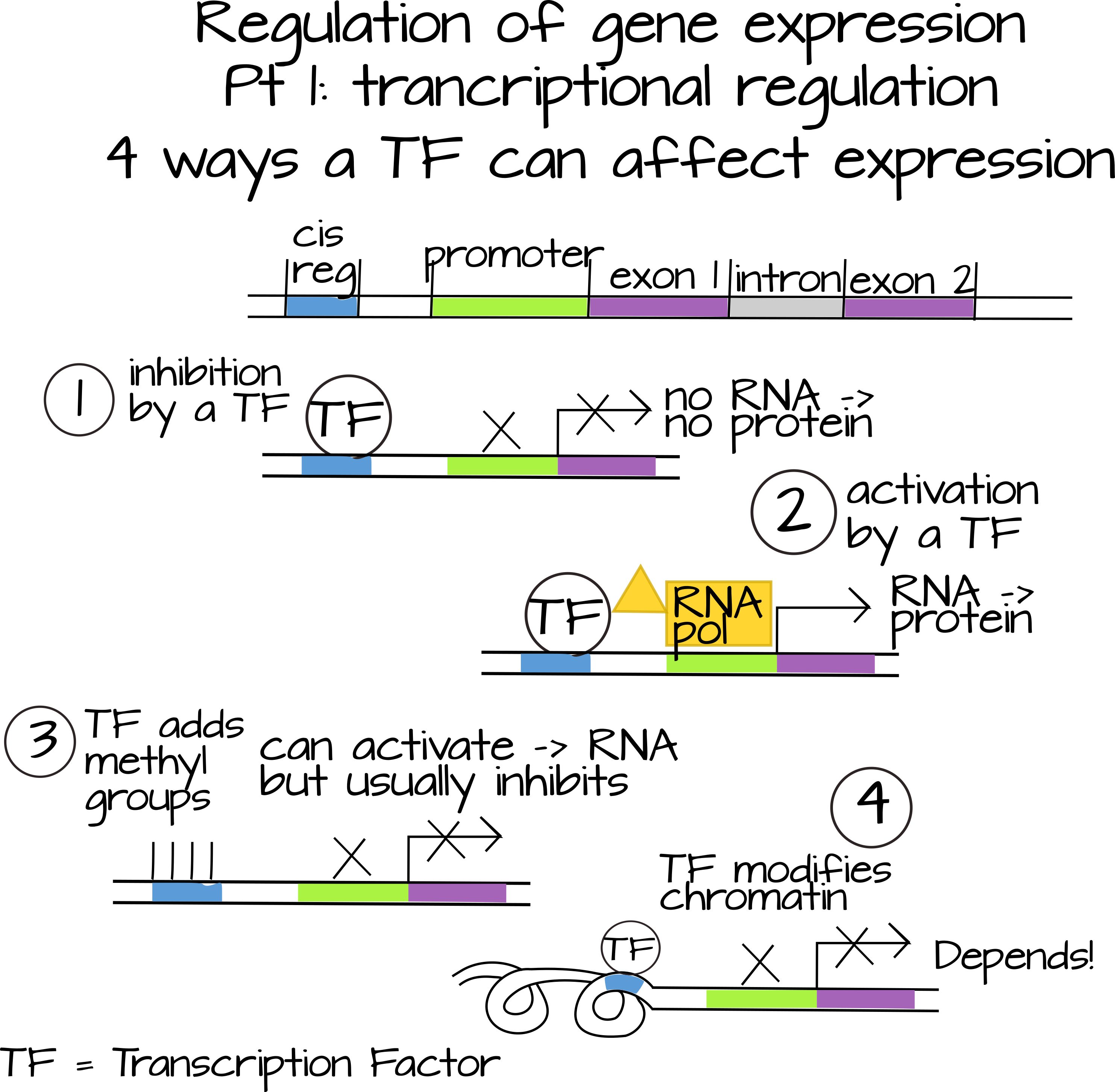
Gene expression gets regulated at three major levels (that can be subdivided into many many sublevels). Transcription itself can be regulated (we often refer to this as turning a gene "on" or "off"). Regulating transcription is the job of transcription factors, also known as "trans acting" factors. These are diffusible molecules, often proteins, that act in many different ways to turn up or down the rate of transcription of a particular gene. Some transcription factors bind directly to to cis-regulatory sequences, pieces of DNA usually near the coding region of a gene, and influence the ability of RNA polymerase to bind to the promoter of the gene. They can do this by directly interacting with proteins involved in RNA polymerase recruitment (for example TFIID and Mediator), or by changing the accessibility of the promoter (for example, physically blocking the promoter, adding or removing methyl groups, or interacting with histones and histone binding proteins). Some transcription factors bind to other transcription factors and affect their ability to bind DNA.
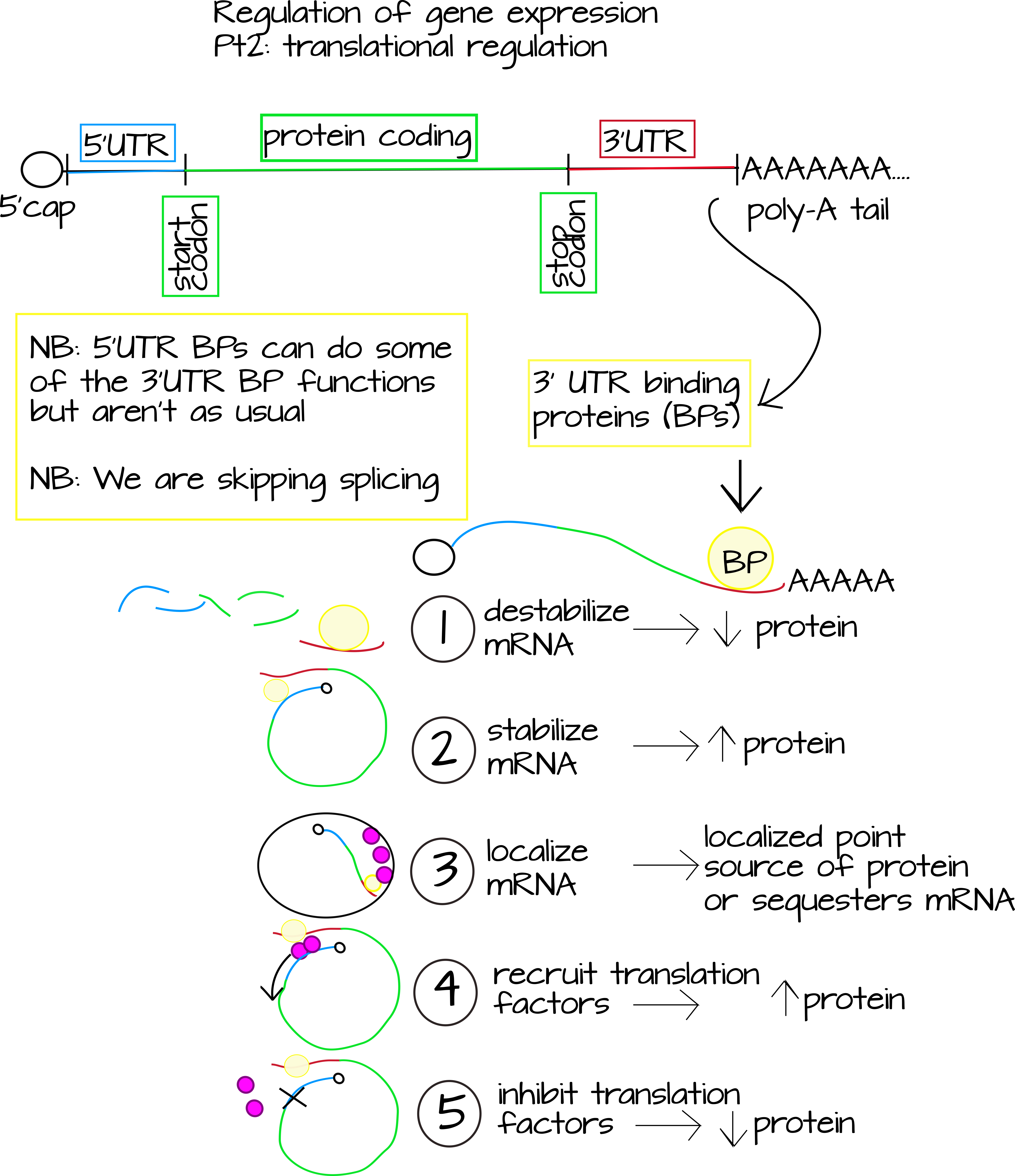
The second major level of gene expression regulation is translational regulation. This includes any kind of regulators that affect the amount of protein/peptide produced by a single mRNA molecule. There are two very well-studied ways that this happens. The first is through RNA interference (RNAi), which you may have heard of. RNAi uses short pieces of non-coding RNA that are exact or very close reverse-complements to part of an mRNA sequence. This targets the mRNA for destruction or blocks translation.1 The second well-studied way to regulate translation is through RNA binding proteins. These proteins will often bind to specific sequences found outside the protein-coding region in mRNAs. These regions are called the untranslated regions or "UTRs". Proteins that bind here can
- Destabilize the mRNA, lowering translation levels.
- Stabilize the mRNA, raising translation levels.
- Localize the RNA to a specific part of the cell.
- Block or inhibit the binding of translation factors, lowering translation levels.
- Recruit translation factors, raising translation levels.2
The third and final major level of regulating gene expression is the post-translational level. This level encompasses most of what we think of as signal transduction pathways, or the protein-protein and protein-small-molecule interactions that affect protein stability and function. This can include both covalent modification of the protein (for example, phosphorylation, dephosphorylation, glycosylation, or proteolytic cleavage) as well as non-covalent modification (for example allosteric inhibition or activation, homo and heterodimer formation).3 If you aren't sure what these terms mean right now, that's ok - most of them we wont cover and those we do cover are easier to understand in context as you learn about signal transduction pathways.
References
- "Mechanisms of gene silencing by double-stranded RNA", 2004, Nature, Gunter Meister & Thomas Tuschl, doi:10.1038/nature02873
- "A brave new world of RNA-binding proteins", 2018, Nature Reviews: Molecular Cell Biology, Matthias W. Hentze, Alfredo Castello, Thomas Schwarzl & Thomas Preiss doi:10.1038/nrm.2017.130
- "Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling", 2002, Genes and Development, Scott Barolo and James W. Posakony, 10.1101/gad.976502


