13.E: Mechanisms of Microbial Genetics (Exercises)
- Page ID
- 78152
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
13.1: Mutations
A mutation is a heritable change in the DNA sequence of an organism. The resulting organism, called a mutant, may have a recognizable change in phenotype compared to the wild type, which is the phenotype most commonly observed in nature. A change in the DNA sequence is conferred to mRNA through transcription, and may lead to an altered amino acid sequence in a protein on translation.
Multiple Choice
Which of the following is a change in the sequence that leads to formation of a stop codon?
- missense mutation
- nonsense mutation
- silent mutation
- deletion mutation
- Answer
-
B
The formation of pyrimidine dimers results from which of the following?
- spontaneous errors by DNA polymerase
- exposure to gamma radiation
- exposure to ultraviolet radiation
- exposure to intercalating agents
- Answer
-
C
Which of the following is an example of a frameshift mutation?
- a deletion of a codon
- missense mutation
- silent mutation
- deletion of one nucleotide
- Answer
-
D
Which of the following is the type of DNA repair in which thymine dimers are directly broken down by the enzyme photolyase?
- direct repair
- nucleotide excision repair
- mismatch repair
- proofreading
- Answer
-
A
Which of the following regarding the Ames test is true?
- It is used to identify newly formed auxotrophic mutants.
- It is used to identify mutants with restored biosynthetic activity.
- It is used to identify spontaneous mutants.
- It is used to identify mutants lacking photoreactivation activity.
- Answer
-
B
Fill in the Blank
A chemical mutagen that is structurally similar to a nucleotide but has different base-pairing rules is called a ________.
- Answer
-
nucleoside analog
The enzyme used in light repair to split thymine dimers is called ________.
- Answer
-
photolyase
The phenotype of an organism that is most commonly observed in nature is called the ________.
- Answer
-
wild type
True/False
Carcinogens are typically mutagenic.
- Answer
-
True
Short Answer
Why is it more likely that insertions or deletions will be more detrimental to a cell than point mutations?
Critical Thinking
Below are several DNA sequences that are mutated compared with the wild-type sequence: 3’-T A C T G A C T G A C G A T C-5’. Envision that each is a section of a DNA molecule that has separated in preparation for transcription, so you are only seeing the template strand. Construct the complementary DNA sequences (indicating 5’ and 3’ ends) for each mutated DNA sequence, then transcribe (indicating 5’ and 3’ ends) the template strands, and translate the mRNA molecules using the genetic code, recording the resulting amino acid sequence (indicating the N and C termini). What type of mutation is each?
|
Mutated DNA Template Strand #1: 3’-T A C T G T C T G A C G A T C-5’ Complementary DNA sequence: mRNA sequence transcribed from template: Amino acid sequence of peptide: Type of mutation: |
|
Mutated DNA Template Strand #2: 3’-T A C G G A C T G A C G A T C-5’ Complementary DNA sequence: mRNA sequence transcribed from template: Amino acid sequence of peptide: Type of mutation: |
|
Mutated DNA Template Strand #3: 3’-T A C T G A C T G A C T A T C-5’ Complementary DNA sequence: mRNA sequence transcribed from template: Amino acid sequence of peptide: Type of mutation: |
|
Mutated DNA Template Strand #4: 3’-T A C G A C T G A C T A T C-5’ Complementary DNA sequence: mRNA sequence transcribed from template: Amino acid sequence of peptide: Type of mutation: |
Why do you think the Ames test is preferable to the use of animal models to screen chemical compounds for mutagenicity?
13.2: How Asexual Prokaryotes Achieve Genetic Diversity
How do organisms whose dominant reproductive mode is asexual create genetic diversity? In prokaryotes, horizontal gene transfer (HGT), the introduction of genetic material from one organism to another organism within the same generation, is an important way to introduce genetic diversity. HGT allows even distantly related species to share genes, influencing their phenotypes.
Multiple Choice
Which is the mechanism by which improper excision of a prophage from a bacterial chromosome results in packaging of bacterial genes near the integration site into a phage head?
- conjugation
- generalized transduction
- specialized transduction
- transformation
- Answer
-
C
Which of the following refers to the uptake of naked DNA from the surrounding environment?
- conjugation
- generalized transduction
- specialized transduction
- transformation
- Answer
-
D
The F plasmid is involved in which of the following processes?
- conjugation
- transduction
- transposition
- transformation
- Answer
-
A
Which of the following refers to the mechanism of horizontal gene transfer naturally responsible for the spread of antibiotic resistance genes within a bacterial population?
- conjugation
- generalized transduction
- specialized transduction
- transformation
- Answer
-
A
Fill in the Blank
A small DNA molecule that has the ability to independently excise from one location in a larger DNA molecule and integrate into the DNA elsewhere is called a ________.
- Answer
-
transposon or transposable element
________ is a group of mechanisms that allow for the introduction of genetic material from one organism to another organism within the same generation.
- Answer
-
Horizontal gene transfer
True/False
Asexually reproducing organisms lack mechanisms for generating genetic diversity within a population.
- Answer
-
False
Short Answer
Briefly describe two ways in which chromosomal DNA from a donor cell may be transferred to a recipient cell during the process of conjugation.
Describe what happens when a nonsense mutation is introduced into the gene encoding transposase within a transposon.
13.3: Gene Regulation - Operon Theory
Genomic DNA contains both structural genes, which encode products that serve as cellular structures or enzymes, and regulatory genes, which encode products that regulate gene expression. The expression of a gene is a highly regulated process. Whereas regulating gene expression in multicellular organisms allows for cellular differentiation, in single-celled organisms like prokaryotes, it ensures that a cell’s resources are not wasted making proteins that the cell does not need at that time.
Multiple Choice
An operon of genes encoding enzymes in a biosynthetic pathway is likely to be which of the following?
- inducible
- repressible
- constitutive
- monocistronic
- Answer
-
B
An operon encoding genes that are transcribed and translated continuously to provide the cell with constant intermediate levels of the protein products is said to be which of the following?
- repressible
- inducible
- constitutive
- activated
- Answer
-
C
Which of the following conditions leads to maximal expression of the lac operon?
- lactose present, glucose absent
- lactose present, glucose present
- lactose absent, glucose absent
- lactose absent, glucose present
- Answer
-
A
Which of the following is a type of regulation of gene expression unique to eukaryotes?
- attenuation
- use of alternate σ factor
- chemical modification of histones
- alarmones
- Answer
-
C
Fill in the Blank
The DNA sequence, to which repressors may bind, that lies between the promoter and the first structural gene is called the ________.
- Answer
-
operator
The prevention of expression of operons encoding substrate use pathways for substrates other than glucose when glucose is present is called _______.
- Answer
-
catabolite repression
Short Answer
What are two ways that bacteria can influence the transcription of multiple different operons simultaneously in response to a particular environmental condition?
Critical Thinking
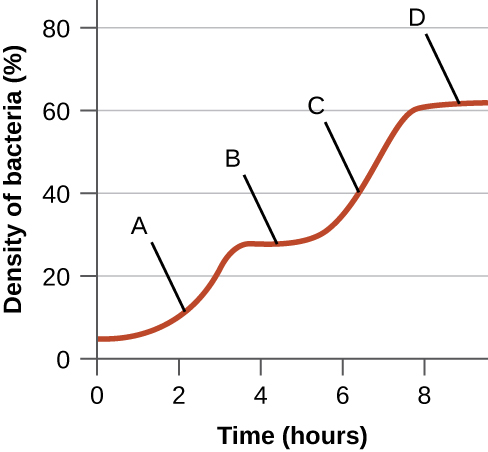
The following figure is from Monod’s original work on diauxic growth showing the growth of E. coli in the simultaneous presence of xylose and glucose as the only carbon sources. Explain what is happening at points A–D with respect to the carbon source being used for growth, and explain whether the xylose-use operon is being expressed (and why). Note that expression of the enzymes required for xylose use is regulated in a manner similar to the expression of the enzymes required for lactose use.