A. The Major Types of Cellular RNA
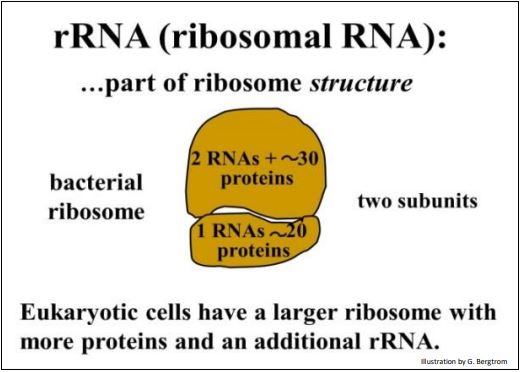
All cells make three main kinds of RNA: ribosomal RNA (rRNA), transfer RNA (tRNA) and messenger RNA (mRNA). rRNA is a structural as well as enzymatic component of ribosomes, the protein-synthesizing machine in the cell. Quantitatively, rRNAs are by far the most abundant RNAs in the cell and mRNAs, the least. Three rRNAs and about 50 ribosomal proteins make up the two subunits of a bacterial ribosome, as illustrated below.
tRNAs are the decoding devices used in protein synthesis (translation) to convert nucleic acid sequence information into the amino acid sequences of polypeptides. tRNAs attached to amino acids are positioned on ribosomes based on codonanticodon recognition, as shown below.

During translation, tRNAs decode base sequences in messenger RNA (mRNAs) into amino acid sequences of polypeptides.
186 Transcription Overview: Ribosomes and Ribosomal RNAs
187 Transcription Overview: Demonstrating the Major RNAs
In 2009, Venkatraman Ramakrishnan, Thomas A. Steitz and ADA Yonath received the Nobel Prize in Chemistry their studies on the structure and molecular biology of the ribosome. Dr. Yonath is one of five women to receive a Nobel Prize – the others were Marie Curie, Irène Joliot-Curie, Dorothy Hodgkin and Barbara Mclintock.
The fact that genes are inside the eukaryotic nucleus and that the synthesis of polypeptides encoded by those genes happens in the cytoplasm led to the proposal that there must be a messenger RNA (mRNA. Sidney Brenner eventually confirmed the existence of mRNAs. Check out his classic experiment in Brenner S (1961, An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190:576-581).
Recall polypeptide synthesis by the formation of polyribosomes (polysomes) along a single mRNA, as illustrated below.
While mRNA is a small fraction of total cellular RNA, there are still smaller amounts of other RNAs such as the transient primers that we saw in DNA replication. We’ll encounter still other kinds of low-abundance RNAs later.
B. Key Steps of Transcription
In transcription, an RNA polymerase uses the template DNA strand of a gene to catalyze synthesis of a complementary, antiparallel RNA strand. RNA polymerases use ribose nucleotide triphosphate (NTP) precursors, in contrast to DNA polymerases, which use deoxyribose nucleotide (dNTP) precursors. In addition, RNAs incorporate uracil (U) nucleotides into RNA strands instead of the thymine (T) nucleotides that end up in new DNA. Another contrast to replication - RNA synthesis does not require a primer. With the help of transcription initiation factors, RNA polymerase locates the transcription start site of a gene and begins synthesis of a new RNA strand from scratch. Finally, like replication, transcription is error-prone.
The basic steps of transcription are summarized on the next page. Here we can identify several of the DNA sequences that characterize a gene. The promoter is the binding site for RNA polymerase. It usually lies 5’ to, or upstream of the transcription start site (the bent arrow). Binding of the RNA polymerase positions the enzyme to near the transcription start site, where it will start unwinding the double helix and begin synthesizing new RNA. The transcribed grey DNA region in each of the three panels are the transcription unit of the gene. Termination sites are typically 3’ to, or downstream from the transcribed region of the gene. By convention, upstream refers to DNA 5’ to a given reference point on the DNA (e.g., the transcription start-site of a gene). Downstream then, refers to DNA 3’ to a given reference point on the DNA.
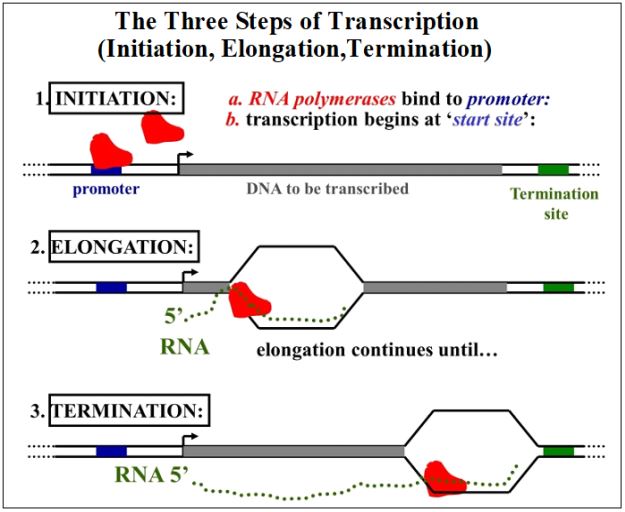
188 Transcription Overview: The Basic Mechanism of RNA Synthesis
In bacteria, some transcription units encode more than one kind of RNA. Bacterial operons are an example of this phenomenon. The resulting mRNAs can be translated into multiple polypeptides at the same time. In the illustration below, RNA polymerase is transcribing a single mRNA molecule encoding three separate polypeptides.

Bacterial transcription of the different RNAs requires only one RNA polymerase. Different RNA polymerases catalyze rRNA, mRNA and tRNA transcription in eukaryotes. Roger Kornberg received the Nobel Prize in Medicine in 2006 for his discovery of the role of RNA polymerase II and other proteins involved in eukaryotic messenger RNA transcription (like father-like son!!).
189 RNA Polymerases in Prokaryotes and Eukaryotes
While mRNAs, rRNAs and tRNAs are most of what cells transcribe, a growing number of other RNAs (e.g., siRNAs, miRNAs, lncRNAs…) are also transcribed. Some functions of these transcripts (including control of gene expression or other transcript use) are discussed elsewhere.
C. RNAs are Extensively Processed After Transcription in Eukaryotes
Many eukaryotic RNAs are processed (trimmed, chemically modified) from large precursor RNAs to mature, functional RNAs. These precursor RNAs (pre-RNAs, or primary transcripts) contain in their sequences the information necessary for their function in the cell.
Processing of the three major types of transcripts in eukaryotes is shown below.
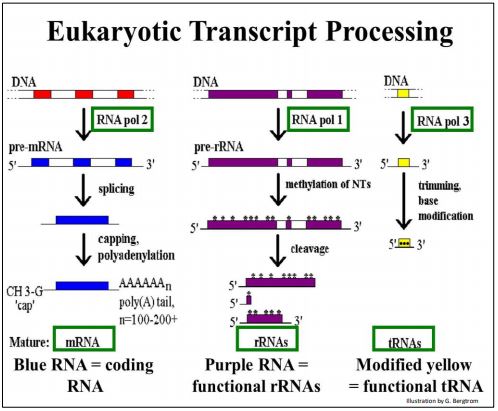
To summarize the illustration:
- Many eukaryotic genes are ‘split’ into coding regions (exons) and non-coding intervening regions (introns).
- Transcription of split genes generates a primary mRNA transcript (pre-mRNA).
- Primary transcripts are spliced to remove the introns from the exons; exons are then ligated into a continuous mRNA. In some cases, the same pre-mRNA is spliced into alternate mRNAs encoding related but not identical polypeptides!
- Pre-rRNA is cleaved and/or trimmed (not spliced!) to make shorter mature rRNAs.
- Pre-tRNAs are trimmed, some bases within the transcript are modified and 3 bases (not encoded by the tRNA gene) are enzymatically added to the 3’-end.
190 Post Transcriptional Processing Overview
The details of transcription and processing differ substantially in prokaryotes and eukaryotes. Let’s focus first on details of transcription itself, and then RNA processing.


