5.5: Mutations
- Page ID
- 75838
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Compare point mutations and frameshift mutations
- Describe the differences between missense, nonsense, and silent mutations
- Explain how different mutagens act
- Compare different types o f repair mechanisms
- Explain why the Ames test can be used to detect carcinogens
- Analyze sequences of DNA and identify examples of types of mutations
A mutation is a heritable change in the DNA sequence of an organism. The resulting organism, called a mutant, may have a recognizable change in phenotype compared to the wild type, which is the phenotype most commonly observed in nature. A change in the DNA sequence is conferred to mRNA through transcription, and may lead to an altered amino acid sequence in a protein on translation. Because proteins carry out the vast majority of cellular functions, a change in amino acid sequence in a protein may lead to an altered phenotype for the cell and organism.
Effects of Mutations on DNA Sequence
There are several types of mutations that are classified according to how the DNA molecule is altered. One type, called a point mutation, affects a single base and most commonly occurs when one base is substituted or replaced by another. Mutations also result from the addition of one or more bases, known as an insertion, or the removal of one or more bases, known as a deletion.
What type of a mutation occurs when a gene has two fewer nucleotides in its sequence?
Effects of Mutations on Protein Structure and Function
Point mutations may have a wide range of effects on protein function (Figure \(\PageIndex{1}\)). As a consequence of the degeneracy of the genetic code, a point mutation will commonly result in the same amino acid being incorporated into the resulting polypeptide despite the sequence change. This change would have no effect on the protein’s structure, and is thus called a silent mutation. A missense mutation results in a different amino acid being incorporated into the resulting polypeptide. The effect of a missense mutation depends on how chemically different the new amino acid is from the wild-type amino acid. The location of the changed amino acid within the protein also is important. For example, if the changed amino acid is part of the enzyme’s active site, then the effect of the missense mutation may be significant. Many missense mutations result in proteins that are still functional, at least to some degree. Sometimes the effects of missense mutations may be only apparent under certain environmental conditions; such missense mutations are called conditional mutations. Rarely, a missense mutation may be beneficial. Under the right environmental conditions, this type of mutation may give the organism that harbors it a selective advantage. Yet another type of point mutation, called a nonsense mutation, converts a codon encoding an amino acid (a sense codon) into a stop codon (a nonsense codon). Nonsense mutations result in the synthesis of proteins that are shorter than the wild type and typically not functional.
Deletions and insertions also cause various effects. Because codons are triplets of nucleotides, insertions or deletions in groups of three nucleotides may lead to the insertion or deletion of one or more amino acids and may not cause significant effects on the resulting protein’s functionality. However, frameshift mutations, caused by insertions or deletions of a number of nucleotides that are not a multiple of three are extremely problematic because a shift in the reading frame results (Figure \(\PageIndex{1}\)). Because ribosomes read the mRNA in triplet codons, frameshift mutations can change every amino acid after the point of the mutation. The new reading frame may also include a stop codon before the end of the coding sequence. Consequently, proteins made from genes containing frameshift mutations are nearly always nonfunctional.
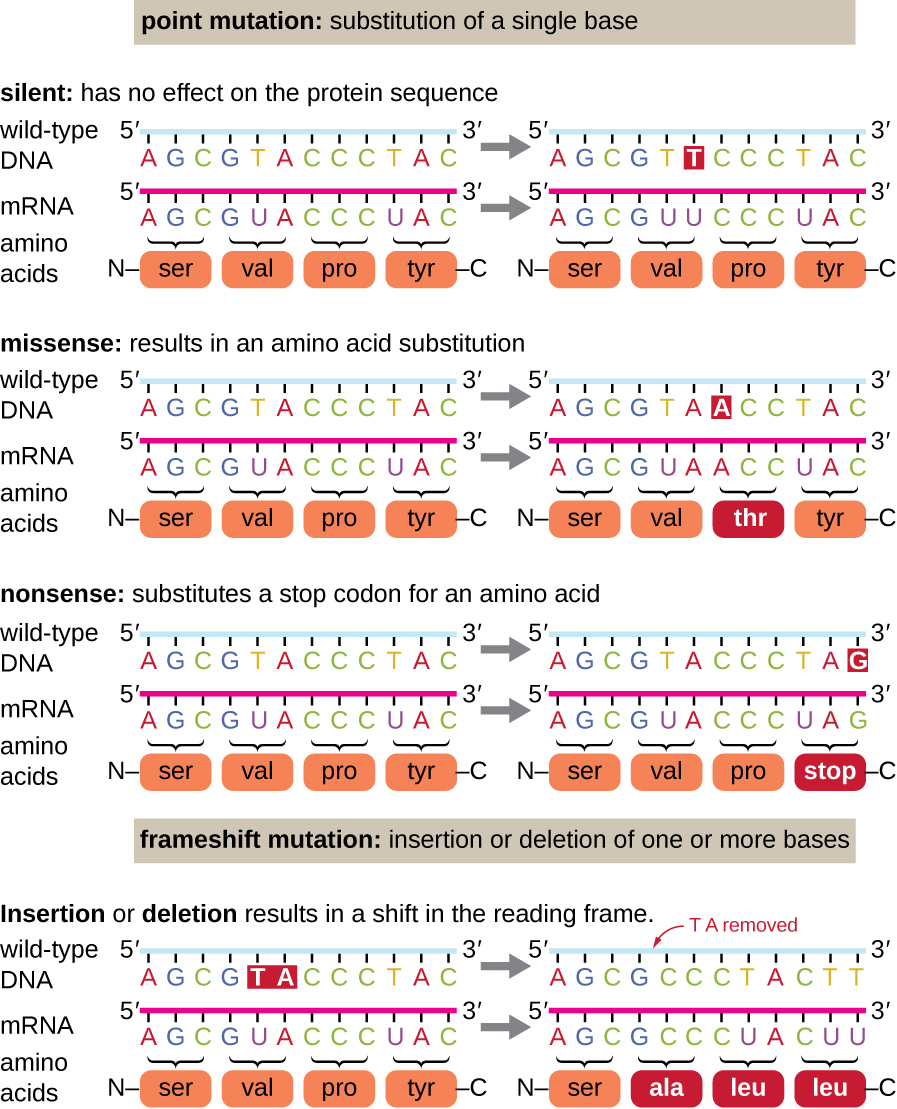
- What are the reasons a nucleotide change in a gene for a protein might not have any effect on the phenotype of that gene?
- Is it possible for an insertion of three nucleotides together after the fifth nucleotide in a protein-coding gene to produce a protein that is shorter than normal? How or how not?
Since the first case of infection with human immunodeficiency virus (HIV) was reported in 1981, nearly 40 million people have died from HIV infection,1 the virus that causes acquired immune deficiency syndrome (AIDS). The virus targets helper T cells that play a key role in bridging the innate and adaptive immune response, infecting and killing cells normally involved in the body’s response to infection. There is no cure for HIV infection, but many drugs have been developed to slow or block the progression of the virus. Although individuals around the world may be infected, the highest prevalence among people 15–49 years old is in sub-Saharan Africa, where nearly one person in 20 is infected, accounting for greater than 70% of the infections worldwide2 (Figure \(\PageIndex{2}\)). Unfortunately, this is also a part of the world where prevention strategies and drugs to treat the infection are the most lacking.
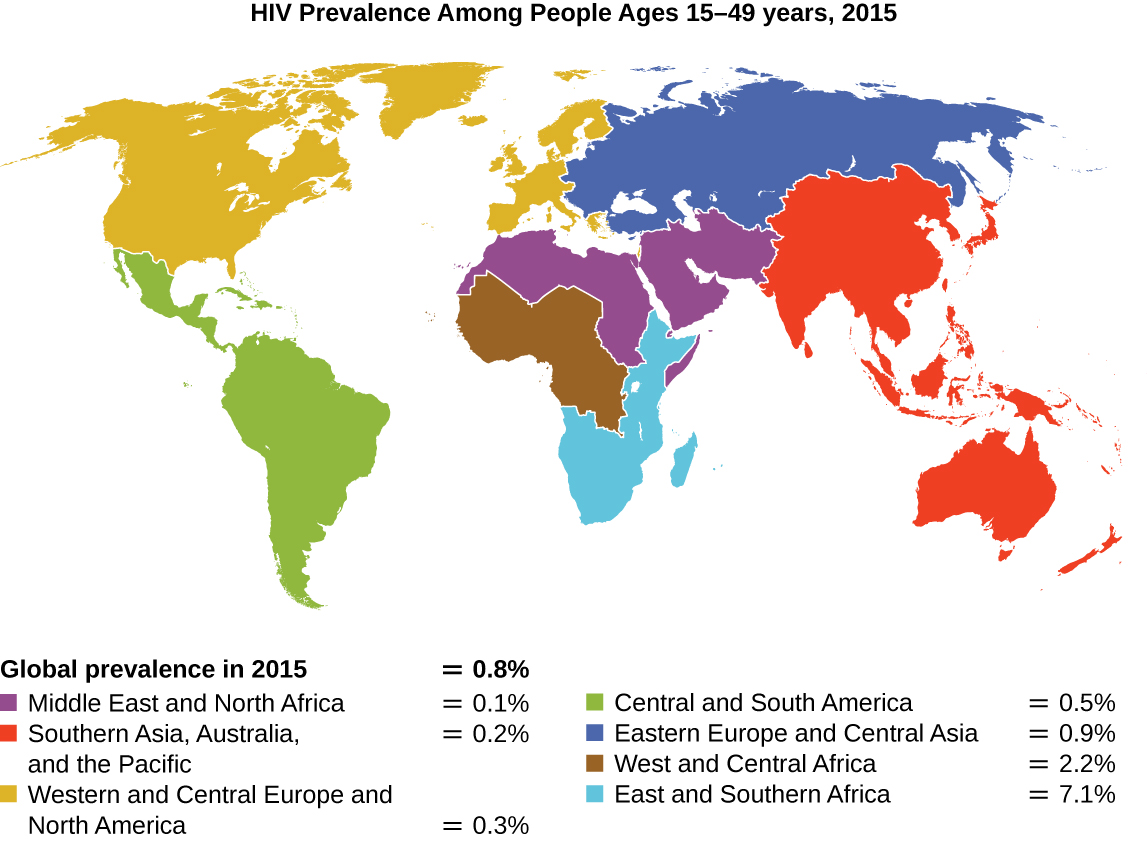
In recent years, scientific interest has been piqued by the discovery of a few individuals from northern Europe who are resistant to HIV infection. In 1998, American geneticist Stephen J. O’Brien at the National Institutes of Health (NIH) and colleagues published the results of their genetic analysis of more than 4,000 individuals. These indicated that many individuals of Eurasian descent (up to 14% in some ethnic groups) have a deletion mutation, called CCR5-delta 32, in the gene encoding CCR5. CCR5 is a coreceptor found on the surface of T cells that is necessary for many strains of the virus to enter the host cell. The mutation leads to the production of a receptor to which HIV cannot effectively bind and thus blocks viral entry. People homozygous for this mutation have greatly reduced susceptibility to HIV infection, and those who are heterozygous have some protection from infection as well.
It is not clear why people of northern European descent, specifically, carry this mutation, but its prevalence seems to be highest in northern Europe and steadily decreases in populations as one moves south. Research indicates that the mutation has been present since before HIV appeared and may have been selected for in European populations as a result of exposure to the plague or smallpox. This mutation may protect individuals from plague (caused by the bacterium Yersinia pestis) and smallpox (caused by the variola virus) because this receptor may also be involved in these diseases. The age of this mutation is a matter of debate, but estimates suggest it appeared between 1875 years to 225 years ago, and may have been spread from Northern Europe through Viking invasions.
This exciting finding has led to new avenues in HIV research, including looking for drugs to block CCR5 binding to HIV in individuals who lack the mutation. Although DNA testing to determine which individuals carry the CCR5-delta 32 mutation is possible, there are documented cases of individuals homozygous for the mutation contracting HIV. For this reason, DNA testing for the mutation is not widely recommended by public health officials so as not to encourage risky behavior in those who carry the mutation. Nevertheless, inhibiting the binding of HIV to CCR5 continues to be a valid strategy for the development of drug therapies for those infected with HIV.
Causes of Mutations
Mistakes in the process of DNA replication can cause spontaneous mutations to occur. The error rate of DNA polymerase is one incorrect base per billion base pairs replicated. Exposure to mutagens can cause induced mutations, which are various types of chemical agents or radiation (Table \(\PageIndex{1}\)). Exposure to a mutagen can increase the rate of mutation more than 1000-fold. Mutagens are often also carcinogens, agents that cause cancer. However, whereas nearly all carcinogens are mutagenic, not all mutagens are necessarily carcinogens.
Chemical Mutagens
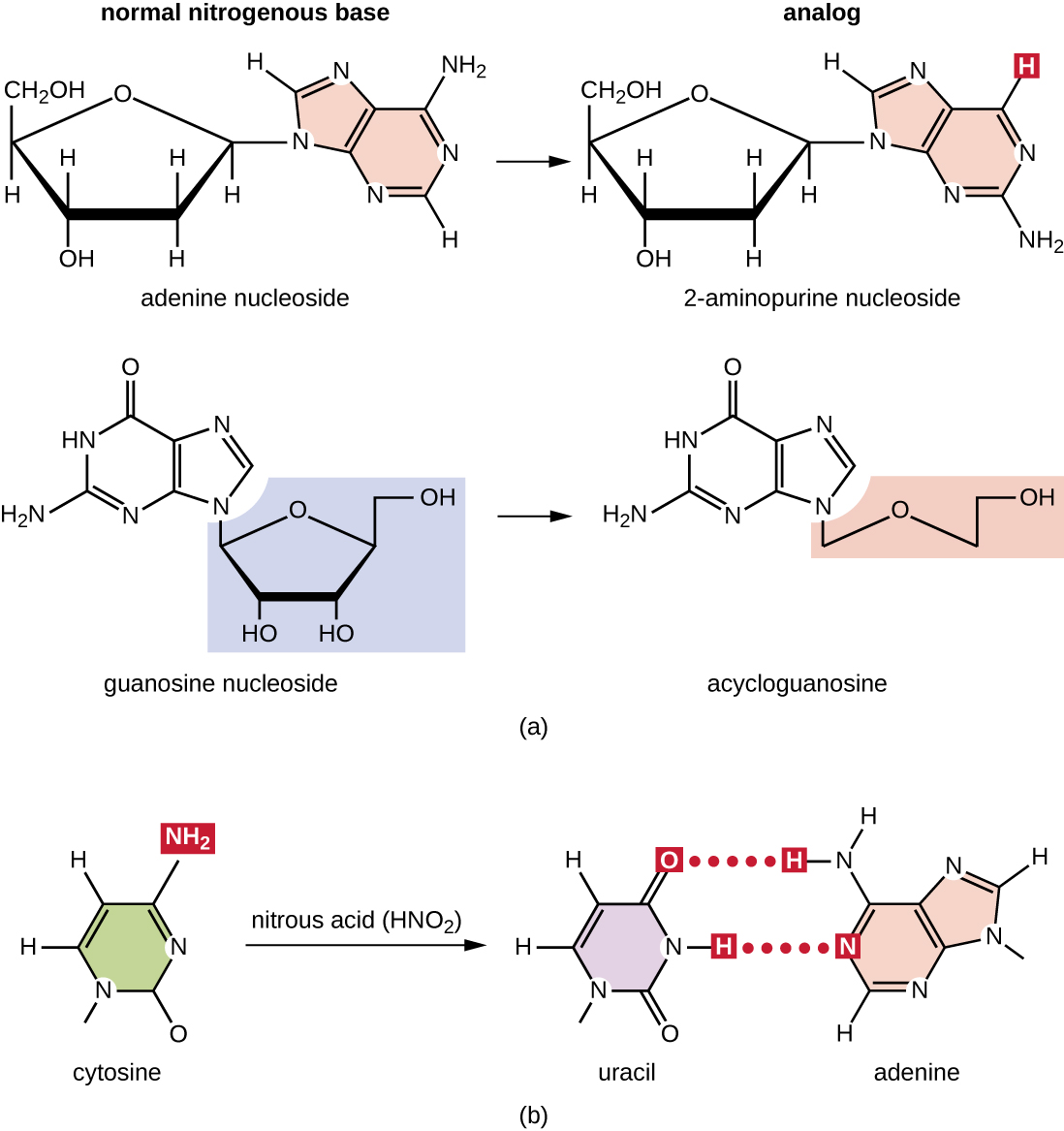
Various types of chemical mutagens interact directly with DNA either by acting as nucleoside analogs or by modifying nucleotide bases. Chemicals called nucleoside analogs are structurally similar to normal nucleotide bases and can be incorporated into DNA during replication (Figure \(\PageIndex{3}\)). These base analogs induce mutations because they often have different base-pairing rules than the bases they replace. Other chemical mutagens can modify normal DNA bases, resulting in different base-pairing rules. For example, nitrous acid deaminates cytosine, converting it to uracil. Uracil then pairs with adenine in a subsequent round of replication, resulting in the conversion of a GC base pair to an AT base pair. Nitrous acid also deaminates adenine to hypoxanthine, which base pairs with cytosine instead of thymine, resulting in the conversion of a TA base pair to a CG base pair.
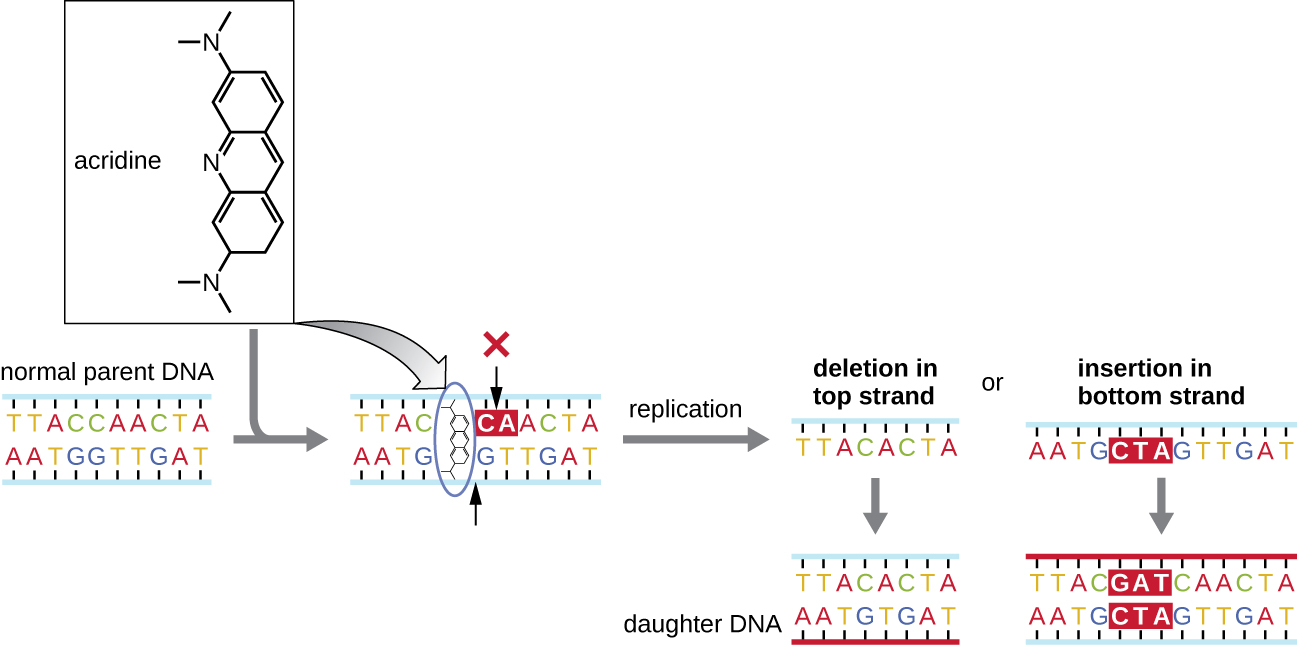
Chemical mutagens known as intercalating agents work differently. These molecules slide between the stacked nitrogenous bases of the DNA double helix, distorting the molecule and creating atypical spacing between nucleotide base pairs (Figure \(\PageIndex{4}\)). As a result, during DNA replication, DNA polymerase may either skip replicating several nucleotides (creating a deletion) or insert extra nucleotides (creating an insertion). Either outcome may lead to a frameshift mutation. Combustion products like polycyclic aromatic hydrocarbons are particularly dangerous intercalating agents that can lead to mutation-caused cancers. The intercalating agents ethidium bromide and acridine orange are commonly used in the laboratory to stain DNA for visualization and are potential mutagens.
Radiation
Exposure to either ionizing or nonionizing radiation can each induce mutations in DNA, although by different mechanisms. Strong ionizing radiation like X-rays and gamma rays can cause single- and double-stranded breaks in the DNA backbone through the formation of hydroxyl radicals on radiation exposure (Figure \(\PageIndex{5}\)). Ionizing radiation can also modify bases; for example, the deamination of cytosine to uracil, analogous to the action of nitrous acid.3 Ionizing radiation exposure is used to kill microbes to sterilize medical devices and foods, because of its dramatic nonspecific effect in damaging DNA, proteins, and other cellular components.
Nonionizing radiation, like ultraviolet light, is not energetic enough to initiate these types of chemical changes. However, nonionizing radiation can induce dimer formation between two adjacent pyrimidine bases, commonly two thymines, within a nucleotide strand. During thymine dimer formation, the two adjacent thymines become covalently linked and, if left unrepaired, both DNA replication and transcription are stalled at this point. DNA polymerase may proceed and replicate the dimer incorrectly, potentially leading to frameshift or point mutations.
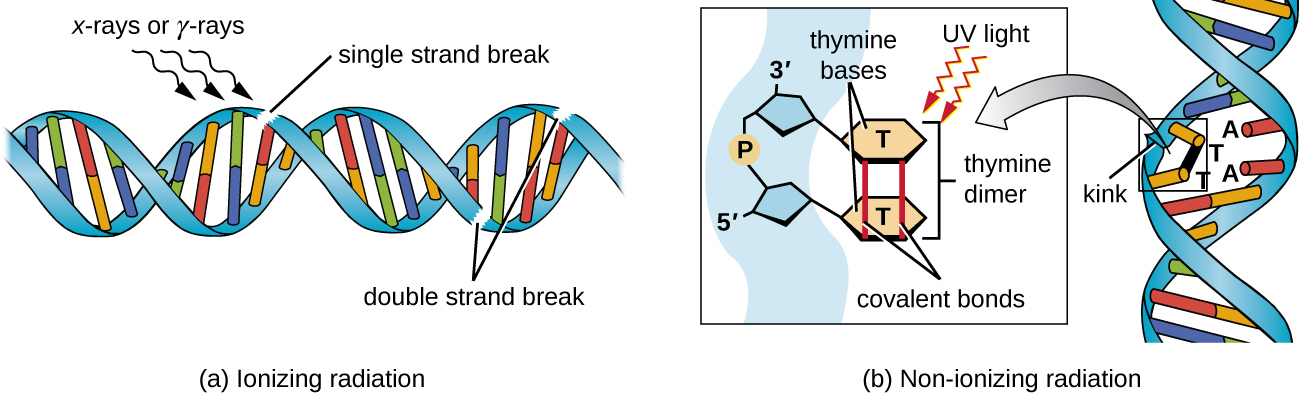
Table \(\PageIndex{1}\): A Summary of Mutagenic Agents
| Mutagenic Agents | Mode of Action | Effect on DNA | Resulting Type of Mutation |
|---|---|---|---|
| Nucleoside analogs | |||
| 2-aminopurine | Is inserted in place of A but base pairs with C | Converts AT to GC base pair | Point |
| 5-bromouracil | Is inserted in place of T but base pairs with G | Converts AT to GC base pair | Point |
| Nucleotide-modifying agent | |||
| Nitrous oxide | Deaminates C to U | Converts GC to AT base pair | Point |
| Intercalating agents | |||
| Acridine orange, ethidium bromide, polycyclic aromatic hydrocarbons | Distorts double helix, creates unusual spacing between nucleotides | Introduces small deletions and insertions | Frameshift |
| Ionizing radiation | |||
| X-rays, γ-rays | Forms hydroxyl radicals | Causes single- and double-strand DNA breaks | Repair mechanisms may introduce mutations |
| X-rays, γ-rays | Modifies bases (e.g., deaminating C to U) | Converts GC to AT base pair | Point |
| Nonionizing radiation | |||
| Ultraviolet | Forms pyrimidine (usually thymine) dimers | Causes DNA replication errors | Frameshift or point |
- How does a base analog introduce a mutation?
- How does an intercalating agent introduce a mutation?
- What type of mutagen causes thymine dimers?
DNA Repair
The process of DNA replication is highly accurate, but mistakes can occur spontaneously or be induced by mutagens. Uncorrected mistakes can lead to serious consequences for the phenotype. Cells have developed several repair mechanisms to minimize the number of mutations that persist.
Proofreading
Most of the mistakes introduced during DNA replication are promptly corrected by most DNA polymerases through a function called proofreading. In proofreading, the DNA polymerase reads the newly added base, ensuring that it is complementary to the corresponding base in the template strand before adding the next one. If an incorrect base has been added, the enzyme makes a cut to release the wrong nucleotide and a new base is added.
Mismatch Repair
Some errors introduced during replication are corrected shortly after the replication machinery has moved. This mechanism is called mismatch repair. The enzymes involved in this mechanism recognize the incorrectly added nucleotide, excise it, and replace it with the correct base. One example is the methyl-directed mismatch repair in E. coli. The DNA is hemimethylated. This means that the parental strand is methylated while the newly synthesized daughter strand is not. It takes several minutes before the new strand is methylated. Proteins MutS, MutL, and MutH bind to the hemimethylated site where the incorrect nucleotide is found. MutH cuts the nonmethylated strand (the new strand). An exonuclease removes a portion of the strand (including the incorrect nucleotide). The gap formed is then filled in by DNA pol III and ligase.
Repair of Thymine Dimers
Because the production of thymine dimers is common (many organisms cannot avoid ultraviolet light), mechanisms have evolved to repair these lesions. In nucleotide excision repair (also called dark repair), enzymes remove the pyrimidine dimer and replace it with the correct nucleotides (Figure \(\PageIndex{6}\)). In E. coli, the DNA is scanned by an enzyme complex. If a distortion in the double helix is found that was introduced by the pyrimidine dimer, the enzyme complex cuts the sugar-phosphate backbone several bases upstream and downstream of the dimer, and the segment of DNA between these two cuts is then enzymatically removed. DNA pol I replaces the missing nucleotides with the correct ones and DNA ligase seals the gap in the sugar-phosphate backbone.
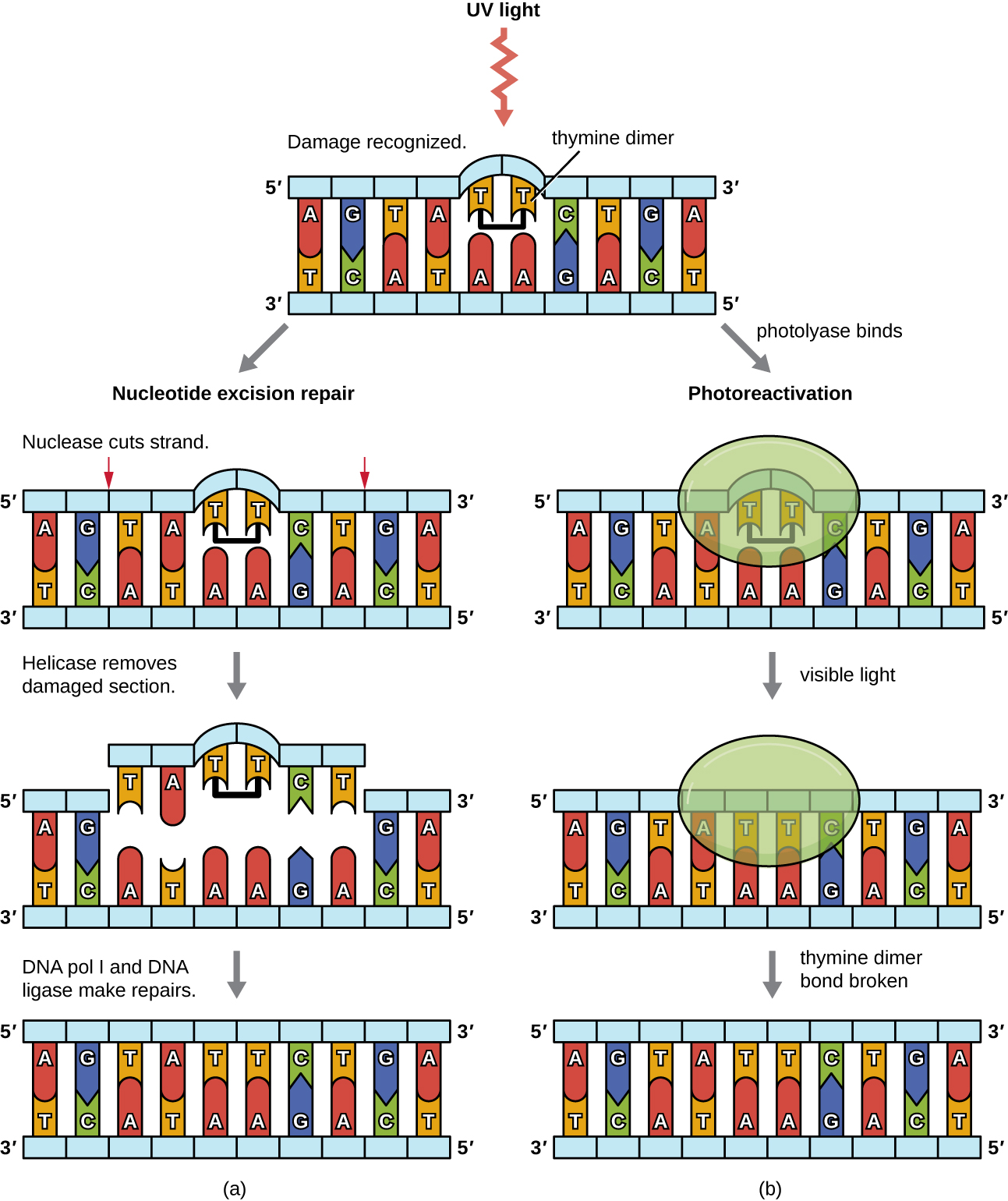
The direct repair (also called light repair) of thymine dimers occurs through the process of photoreactivation in the presence of visible light. An enzyme called photolyase recognizes the distortion in the DNA helix caused by the thymine dimer and binds to the dimer. Then, in the presence of visible light, the photolyase enzyme changes conformation and breaks apart the thymine dimer, allowing the thymines to again correctly base pair with the adenines on the complementary strand. Photoreactivation appears to be present in all organisms, with the exception of placental mammals, including humans. Photoreactivation is particularly important for organisms chronically exposed to ultraviolet radiation, like plants, photosynthetic bacteria, algae, and corals, to prevent the accumulation of mutations caused by thymine dimer formation
- During mismatch repair, how does the enzyme recognize which is the new and which is the old strand?
- How does an intercalating agent introduce a mutation?
- What type of mutation does photolyase repair?
Identifying Bacterial Mutants
One common technique used to identify bacterial mutants is called replica plating. This technique is used to detect nutritional mutants, called auxotrophs, which have a mutation in a gene encoding an enzyme in the biosynthesis pathway of a specific nutrient, such as an amino acid. As a result, whereas wild-type cells retain the ability to grow normally on a medium lacking the specific nutrient, auxotrophs are unable to grow on such a medium. During replica plating (Figure \(\PageIndex{7}\)), a population of bacterial cells is mutagenized and then plated as individual cells on a complex nutritionally complete plate and allowed to grow into colonies. Cells from these colonies are removed from this master plate, often using sterile velvet. This velvet, containing cells, is then pressed in the same orientation onto plates of various media. At least one plate should also be nutritionally complete to ensure that cells are being properly transferred between the plates. The other plates lack specific nutrients, allowing the researcher to discover various auxotrophic mutants unable to produce specific nutrients. Cells from the corresponding colony on the nutritionally complete plate can be used to recover the mutant for further study.
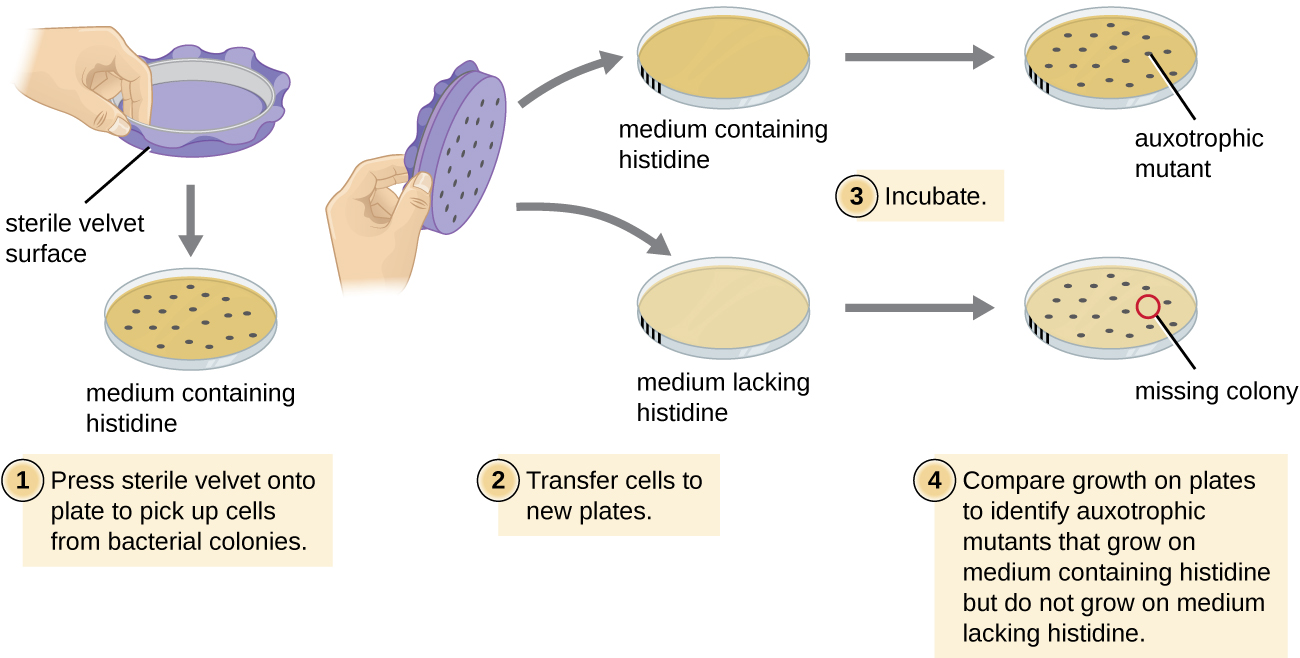
Why are cells plated on a nutritionally complete plate in addition to nutrient-deficient plates when looking for a mutant?
The Ames Test
The Ames test, developed by Bruce Ames (1928–) in the 1970s, is a method that uses bacteria for rapid, inexpensive screening of the carcinogenic potential of new chemical compounds. The test measures the mutation rate associated with exposure to the compound, which, if elevated, may indicate that exposure to this compound is associated with greater cancer risk. The Ames test uses as the test organism a strain of Salmonella typhimurium that is a histidine auxotroph, unable to synthesize its own histidine because of a mutation in an essential gene required for its synthesis. After exposure to a potential mutagen, these bacteria are plated onto a medium lacking histidine, and the number of mutants regaining the ability to synthesize histidine is recorded and compared with the number of such mutants that arise in the absence of the potential mutagen (Figure \(\PageIndex{8}\)). Chemicals that are more mutagenic will bring about more mutants with restored histidine synthesis in the Ames test. Because many chemicals are not directly mutagenic but are metabolized to mutagenic forms by liver enzymes, rat liver extract is commonly included at the start of this experiment to mimic liver metabolism. After the Ames test is conducted, compounds identified as mutagenic are further tested for their potential carcinogenic properties by using other models, including animal models like mice and rats.
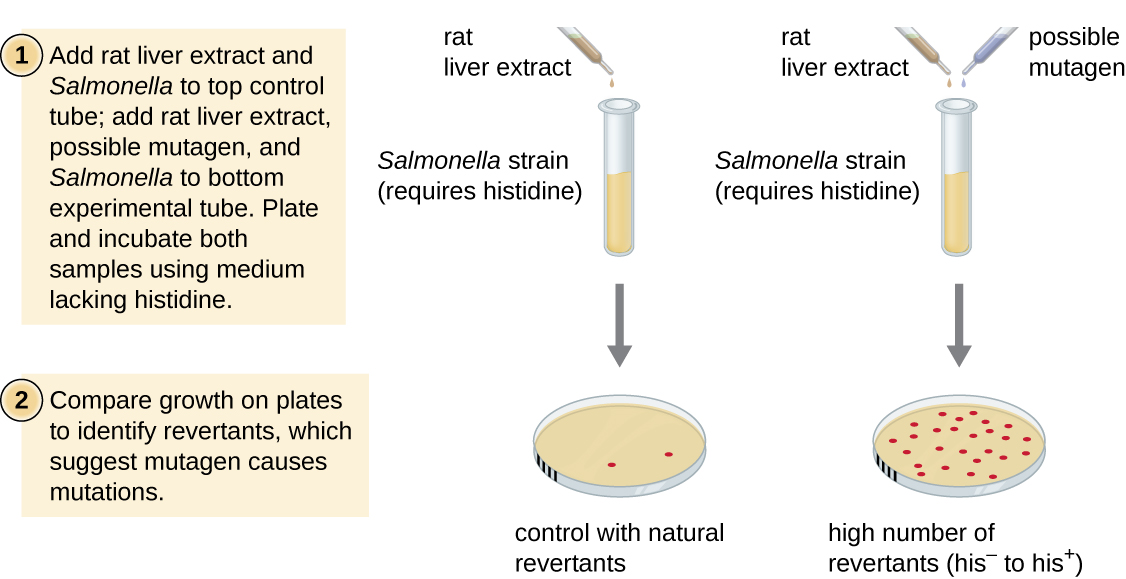
- What mutation is used as an indicator of mutation rate in the Ames test?
- Why can the Ames test work as a test for carcinogenicity?
Key Concepts and Summary
- A mutation is a heritable change in DNA. A mutation may lead to a change in the amino-acid sequence of a protein, possibly affecting its function.
- A point mutation affects a single base pair. A point mutation may cause a silent mutation if the mRNA codon codes for the same amino acid, a missense mutation if the mRNA codon codes for a different amino acid, or a nonsense mutation if the mRNA codon becomes a stop codon.
- Missense mutations may retain function, depending on the chemistry of the new amino acid and its location in the protein. Nonsense mutations produce truncated and frequently nonfunctional proteins.
- A frameshift mutation results from an insertion or deletion of a number of nucleotides that is not a multiple of three. The change in reading frame alters every amino acid after the point of the mutation and results in a nonfunctional protein.
- Spontaneous mutations occur through DNA replication errors, whereas induced mutations occur through exposure to a mutagen.
- Mutagenic agents are frequently carcinogenic but not always. However, nearly all carcinogens are mutagenic.
- Chemical mutagens include base analogs and chemicals that modify existing bases. In both cases, mutations are introduced after several rounds of DNA replication.
- Ionizing radiation, such as X-rays and γ-rays, leads to breakage of the phosphodiester backbone of DNA and can also chemically modify bases to alter their base-pairing rules.
- Nonionizing radiation like ultraviolet light may introduce pyrimidine (thymine) dimers, which, during DNA replication and transcription, may introduce frameshift or point mutations.
- Cells have mechanisms to repair naturally occurring mutations. DNA polymerase has proofreading activity. Mismatch repair is a process to repair incorrectly incorporated bases after DNA replication has been completed.
- Pyrimidine dimers can also be repaired. In nucleotide excision repair (dark repair), enzymes recognize the distortion introduced by the pyrimidine dimer and replace the damaged strand with the correct bases, using the undamaged DNA strand as a template. Bacteria and other organisms may also use direct repair, in which the photolyase enzyme, in the presence of visible light, breaks apart the pyrimidines.
- Through comparison of growth on the complete plate and lack of growth on media lacking specific nutrients, specific loss-of-function mutants called auxotrophs can be identified.
- The Ames test is an inexpensive method that uses auxotrophic bacteria to measure mutagenicity of a chemical compound. Mutagenicity is an indicator of carcinogenic potential.
Footnotes
- 1 World Health Organization. “ Global Health Observatory (GHO) Data, HIV/AIDS.” http://www.who.int/gho/hiv/en/. Accessed August 5, 2016.
- 2 World Health Organization. “ Global Health Observatory (GHO) Data, HIV/AIDS.” http://www.who.int/gho/hiv/en/. Accessed August 5, 2016.
- 3 K.R. Tindall et al. “Changes in DNA Base Sequence Induced by Gamma-Ray Mutagenesis of Lambda Phage and Prophage.” Genetics 118 no. 4 (1988):551–560.
Contributors and Attributions
Nina Parker, (Shenandoah University), Mark Schneegurt (Wichita State University), Anh-Hue Thi Tu (Georgia Southwestern State University), Philip Lister (Central New Mexico Community College), and Brian M. Forster (Saint Joseph’s University) with many contributing authors. Original content via Openstax (CC BY 4.0; Access for free at https://openstax.org/books/microbiology/pages/1-introduction)


