12: Completion of the ID Project
- Page ID
- 110868
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)By the end of this lab period, you will be able to
- Articulate the value of confirmation testing
- Describe the difference between alpha, beta, and gamma hemolysis
- Explain the utility of MSA, bile esculin, EMB, and Hektoen media.
- Finalize the identification of your Gram-positive and your Gram-negative unknowns.
Introduction
At this stage of the ID project, you should have a pretty good idea which unknowns you have, or have narrowed down your Gram-positive and Gram-negative organisms to a few final possibilities. In this lab, you will have the opportunity to perform a few more tests that can help you confirm or exclude those possibilities. All of the tests from today’s lab must go into your test table, and be described BUT some of these tests are only completed IF you believe you have certain organisms.
The tests that we will be setting up today are
- Sheep’s Blood Agar - everyone should perform this test on their Gram-positive organism.
- Mannitol Salt Agar (MSA) - this test can help confirm that you have Staphylococcus aureus or epidermidis. It can be useful in differentiating between these two organisms and other Gram-positive cocci.
- Bile esculin - this test can help confirm the presence of Enterococcus or Klebsiella organisms.
- Eosin Methylene Blue (EMB) - this media can help confirm the presence of a strong fermenter. In particular, this test can be useful to confirm E. coli.
- Hektoen Enteric Agar - this media is useful for confirming the presence of organisms that produce hydrogen sulfide gas, such as Salmonella, and differentiating it from other non-fermenters that are not H2S producers.
- Spore-stain - this technique that we learned about in Lab #7, is important for confirming a Bacillus species and can be used to differentiate Bacillus from other Gram-positive rods.
-
Sheep’s Blood Agar
The ability to break down red blood cells (hemolysis) is a virulence factor characteristic of many organisms including Group A Streptococcus. These bacteria secrete enzymes called “hemolysins”. By breaking down the red blood cells, bacteria are able to release and use the nutrients present in the cells, including heme.
Hemolysis is divided into 3 categories, 𝛂 (alpha), 𝞫 (beta), and 𝛄 (gamma).
- 𝞫-hemolytic organisms break down blood cells completely. They are identified by growing on a blood agar plate, and then noting a complete clearing of blood from around the bacterial growth.
- 𝛂-hemolytic organisms partially break down blood cells. Clearing is not observed, but the agar around the bacteria can appear greenish as a result of this partial breakdown.
- 𝛄-hemolytic organisms are NOT hemolytic. No breakdown of RBCs is observed.

Mannitol Salt Agar
Mannitol Salt Agar (MSA) is a highly selective medium that contains 7.5% sodium chloride. This level of salt inhibits the growth of most bacteria, but organisms found on your (salty) skin, are adapted to higher salt, and are able to grow in this medium. This medium is excellent for confirming Staphylococcus (or ruling it out, if you have any other Gram-positive cocci).
You should know already whether or not your organism ferments mannitol from your phenol red broths. However, MSA is also differential for mannitol fermentation. The medium contains both mannitol as well as the pH indicator phenol red. Recall that phenol red turns yellow below pH 6.8. Organisms that both grow on MSA (salt tolerant) and ferment mannitol will turn the medium a bright yellow color.
.png?revision=1)
Bile Esculin Agar
Bile esculin agar can be used to confirm both Enterococcus and Klebsiella species, but it’s frequently used to differentiate Enterococcus from Streptococcus. Bile esculin agar is selective for Gram-negative bacteria due to the bile, but Gram-positive Enterococcus and Group D Streptococcus species can still grow on this medium. Esculin is a glucoside that is found in some plants, and glucose is a product of its hydrolysis. The other byproduct of hydrolysis is a molecule called esculetin, which reacts with iron (Fe3+) in the medium to form a dark brown precipitate.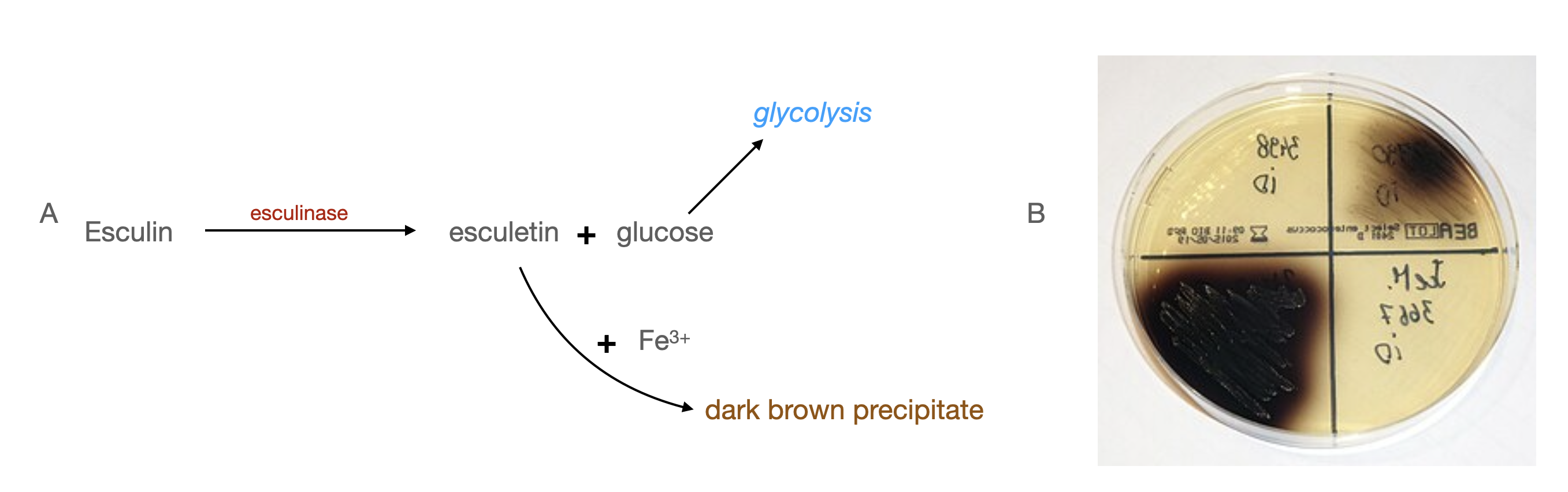
Figure \(\PageIndex{3}\): A. Esculin can be used by bacteria that make the enzyme esculinase to generate glucose for energy. A bile esculin plate showing the brown precipitate formed when Enterococcus is grown on the medium. Stefan Walkowski, CC BY-SA 4.0, via Wikimedia Commons
Eosin-Methylene Blue Agar
EMB (eosin-methylene blue) agar is frequently used to identify fecal coliform bacteria and is both selective and differential. It can be very useful in distinguishing a vigorous fermenter such as E. coli, from other fermenters. The dyes present in the medium including Eosin Y and methylene blue, inhibit the growth of Gram-positive bacteria. These dyes are also pH indicators that will form a dark purple precipitate at low pH. EMB plates also contain both sucrose and lactose. Organisms that ferment these sugars, and lower the pH, will turn purple on this medium. However, when the pH drops very low, below 4.9, the Eosin Y and methylene blue will form a complex with a green color. Very vigorous fermenters, such as E. col, will form colonies with a green metallic sheen.
We will be using EMB agar again during our Food Microbiology lab.
.png?revision=1)
Hektoen Enteric Agar
Hektoen enteric agar is both selective and differential and was designed primarily to differentiate between Salmonella and Shigella in human specimens. It contains bile salts, bromothymol blue, and acid fuchsin, all of which select against the growth of Gram-positive bacteria. It also contains Ferric ammonium citrate or thiosulfate, which will react with hydrogen sulfide (H2S) to form a black precipitate - similar in principle to the reaction that occurred in the SIMs tubes in Lab #11.
Hektoen also contains lactose, sucrose, and salicin. Organisms such as E. coli will ferment these sugars producing acid, which will turn the Bromothymol blue an orange or salmon color. In theory, neither Salmonella nor Shigella will ferment these sugars, and in fact, they don’t in our phenol red broths. However, we often see Shigella turning orange on Hektoen in our lab, so don’t use an orange colony on Hektoen to rule out Shigella.
We will be using Hektoen agar again during our Food Microbiology lab.
.png?revision=1)
Spore Stains
Gram-positive organisms in the Bacillus genera are spore formers. If you believe you have a Bacillus as your Gram-positive, or if you have a Gram-positive rod, you should perform a spore stain to confirm or rule out Bacillus.
To review spore stains and access the staining protocol, please refer to Lab #7.
Materials
- One sheep’s blood agar plate
- One MSA (mannitol salt agar) plate if confirming or ruling out Staphylococcus
- One bile esculin plate if confirming or ruling out Enterococcus or Klebsiella
- One EMB(eosin methylene blue) plate if confirming E. coli, or other strong fermenter.
- One Hektoen enteric agar plate if confirming an H2S producer
- Spore staining materials if confirming or ruling out Bacillus.
Experiment
Day 1
- Perform a streak plate on sheep’s blood agar.
- If you are confirming Enterococcus or Klebsiella, streak out a bile esculin plate as needed.
- If your Gram-negative bacterium is an aggressive fermenter such as E. coli, streak it out on EMB to confirm or rule it out.
- If you suspect your Gram-negative organism to be Salmonella or Shigella, the HE agar plate is a good confirmation test. Additionally, if there is any ambiguity in the lactose-fermentation status of your Gram-negative bacterium (your MacConkey plate and Lactose phenol red test gave contradictory results), use the HE agar to verify whether you have a coliform or not. Note that any H2S producer will produce black colonies on Hektoen.
- If you want to confirm or rule out Staphylococcus, streak it out on MSA.
- If you need to confirm or rule out Bacillus, be sure to perform a spore stain (I will be looking for this when I grade your projects if you report Bacillus!).
- If you need to repeat any tests from prior weeks (Lab #10 or Lab #11), you should set those up today.
- Incubate all newly inoculated media at 37oC for 48 hours or 30oC for 5 days.
-
Day 2
- Observe the hemolysis status of your bacteria from the blood agar plate. Hold the plate up to the light, or observe it over a light box to determine if there are zones of clearing. It is difficult to determine whether or not an organism is alpha or gamma hemolytic. We will record this as a “yes-no” result - your organism is either beta-hemolytic or it’s not.
- Observe any results from EMB, MSA, bile esculin, or Hektoen plates that you may have prepared.
- If you need to confirm or rule out Bacillus, be sure to perform a spore stain if you have not done so already.
- If you need to repeat any tests from prior weeks (Lab #10 or Lab #11), you should set those up today, as this will probably be your last chance.
- Incubate all newly inoculated media at 37oC for 48 hours or 30oC for 5 days.
Data
Be sure to photograph and record all of your results from this week’s tests and include all of them in your test table. Even if you didn’t perform the test, all of them are required to be recorded in your table. Any test not performed can be explained - for example, you might say that you didn’t perform a spore stain because your Gram-positive organism was a cocci.

