1.11: Unknown Lab
- Page ID
- 132783
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Learning Objectives
- Identify the species of an unknown bacterial isolate.
Your unknown strain can be your environmental isolate if it is Gram negative. If not, you can use a lab unknown.
Introduction
We know how to isolate bacteria from the environment and how to select out certain species while not growing others (using selective and differential media). Bacteria are divided into species just like higher organisms. Members of the same species can still vary considerably; these are called strains. Consider the case of E. coli: some strains of E. coli may cause hemolytic uremia (destruction of red blood cells), some cause urinary tract infections, while another strain may be a harmless resident of your intestines. If these strains were on petri plates, they would all look similar. How would you tell these strains apart in the lab?
One way to determine the similarity of strains (called typing) is to test the biochemical reactions the bacteria can perform. Most of the biochemical tests are similar to those in differential media: consumption of a molecule changes the pH, leading to a colour change from a pH indicator dye.
When reading the tests, remember to distinguish between no growth and no reaction. A strain that cannot grow in a medium because it lacks an essential nutrient is much different from a strain that can grow but produces a negative reaction for the biochemical test.
In the microbiology laboratory, samples are often received and it the job of the technologist to identify the organisms in the sample. Typically, bacteria are identified by culture-based approaches (using selective and differential media), microscopy (e.g. Gram staining), and molecular approaches. In this lab, you will identify an unknown organism using microscopy and culture.
Many important human and plant pathogens are in the Enterobacteriaceae family. This family has four main traits:
- Gram negative
- Ferment glucose
- Test negative for oxidase
- Possess the enzyme catalase
Your Gram negative unknown may belong to the Enterobacteriaceae or be a non-Enterobacteriaceae. The non-Enterobacteriaceae group has three main traits:
- Gram negative
- Do not ferment glucose, but may oxidize it
- Many are oxidase positive
This lab is a summary of all the techniques you have learned to date. In addition, we will use a few new tests:
Gelatin hydrolysis
This medium is broth solidified with gelatin instead of agar. If the bacteria possess the enzyme gelatinase, they will liquefy the medium. Gelatinases are secreted proteases that degrade the protein gelatin. Since only some bacteria possess this enzyme, it is a useful test to differentiate closely related species; Serratia, Pseudomonas, Bacillus, and Proteus are positive for gelatinase production.
SIM medium (Sulfide-indole-motility)
This is a multi-test medium. It is a semi-solid agar in a test tube. Sulfide production is detected by the presence of a black precipitate (Fig. 11.1 Tube C), indole is detected by the same method as done previously using TSB and Kovac’s reagent (Fig 11.1 Tube D), and motility is detected as haziness emerging from the stab line (Fig. 11.1 Tube A is motile and Tube B is non-motile).

Oxidase Test
This tests the presence of cytochrome c, a component of the electron transport chain. When cytochrome c is present, the dye is oxidized, it turns from colourless to purple. Since only some bacteria possess cytochrome c, this is a useful test to differentiate bacteria.
Methyl Red and Voges-Proskauer tests
The methyl red test detects the ability of bacteria to ferment sugars by the mixed acid pathway. This results in a greater amount of acidic end products (as carbon is diverted into acids through fermentation). By adding the pH indicator methyl red after growth in MR-VP broth, a red colour indicates a pH below 4.4 ( Fig. 11.2 Tube D, positive for methyl red) or yellow if the pH is higher (Fig 11.2 Tube C, negative for methyl red).
The Voges-Proskauer test detects the production of acetoin, an intermediate in pyruvate fermentation, into 2,3-butanediol. A red colour after the addition of Barrit’s A and B reagents indicates the presence of acetion, a positive reaction (Fig 11.2 Tube B). A yellow colour indicates no acetoin was produced (Fig 11.2 Tube A).

Catalase Test
Catalase is an enzyme produced by some bacteria to prevent damage due to hydrogen peroxide (a by-product of aerobic metabolism). If catalase is produced, hydrogen peroxide will be degraded into water and oxygen. Visible bubbling around the colony indicates catalase is working.
Carbohydrate Fermentation (Phenol Red Glucose, Sucrose or Lactose)
This media has one carbohydrate as the sole carbon source and phenol red, a pH indicator. If the bacteria consume the carbohydrate to produce acids (fermentation), phenol red turns from orange-red to yellow. If the bacteria consume the carbohydrate without fermentation, the medium does not turn yellow but will be turbid.
This medium is a broth and is performed in a test tube. Inside the test tube is an inverted miniature test tube called a Durham tube. If the bacteria produce gas, it will be captured by the Durham tube.
The results for this test could be the following:
- NG (no growth) (Figure 11.3, Tube A
- negative for consumption of the carbohydrate, but the strain grew (Fig. 11.3 Tube C)
- positive for consumption of the carbohydrate and no gas production
- positive for consumption of the carbohydrate with gas production (Fig. 11.3 Tube B)

Citrate Utilization (Simmon’s Citrate)
In this medium, citrate is the sole carbon source. If the bacteria possess citrate permease (a transporter to import citrate), they can grow on this medium. Simmon’s citrate also contains ammonium phosphate as the sole nitrogen source. As the bacteria grow, this is converted into ammonia and ammonium hydroxide, both of which alkalinize the agar. Bromothymol blue is the indicator; as the pH increases, it turns from green at pH 6.9 to blue at pH 7.6.
For this test, growth is linked to the reaction. An organism that grows and consumes citrate will also raise the pH of the agar (Fig. 11.4 Tube B). Strains that are unable to use citrate will be unable to grow (Fig. 11.4 Tube A).
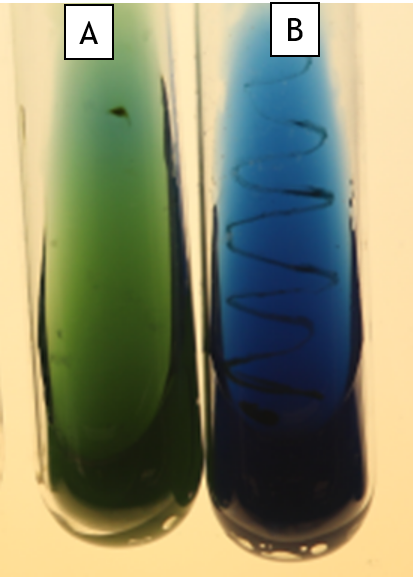
Decarboxylase of Amino Acids (Lysine Decarboxylase)
This medium tests the ability of bacteria to decarboxylate an amino acid (remove the COOH-). It is used to differentiate enterics from other bacteria. The pH indicator, bromocresol purple, is purple at pH 6.8 and above and yellow at pH 5.2 and below (Fig. 11.5 Tube A shows an uninoculated tube). Degradation of amino acids increases the pH of the medium, causing it to turn purple (Fig 11.5. Tube B and Tube C show a weak positive). If the bacteria do not degrade the amino acid but they make acid from glucose, then the tube will turn yellow (Fig 11.5, Tube D)
This test must be conducted anaerobically to cause expression of the decarboxylase enzyme; thus mineral oil is overlaid on the tubes.

Indole production (tryptone broth)
The ability of bacteria to produce indole from tryptophan is a way of differentiating closely related strains. By including tryptophan in the growth medium, indole is produced if the organism is able to. SIM media also contains tryptophan, to allow the indole test to be completed.
Indole is detected after growth by adding Kovac’s reagent (containing dimethylaminobenzaldehyde (DMABA) and HCl in alcohol). The DMABA reacts with any indole present to produce a red colour.
Nitrate Reduction
This tests the ability of a strain to reduce nitrate to nitrite or nitrate to nitrogen gas. Nitrate respiration is a form of anaerobic respiration, with nitrate used in place of oxygen as the terminal electron acceptor. When nitrate accepts electrons, it becomes nitrite. If the bacteria possess the correct enzymes, nitrite can then be reduced into nitrogen gas.
This is a broth medium, and the test tube contains an inverted Durham tube. After growth, nitrate A and B (sulfanilic acid and a-naphthylamine) are added to the tube. If nitrite is present, the medium will turn red due to nitrite reacting with the reagents (Fig. 11.6 Tube A). If the nitrite has been reduced to nitrogen gas OR there is no nitrite present, the medium remains colourless (Fig. 11.6 Tube B). If this is the case, powdered zinc is added. The zinc will reduce any remaining nitrate into nitrite, which then reacts with nitrate A and B, causing the red colour (Fig. 11.6 Tube D). Thus, a red colour after the addition of zinc is negative for nitrate reduction, while a colourless result indicates nitrate was reduced to nitrogen gas or other nitrogenous compounds (Fig. 11.6 Tube C). Gas in the Durham tube indicates the presence of nitrogen gas.

Urease Production
This differentiates organisms based on their ability to degrade urea. If bacteria possess urease, urea is converted into ammonia and carbon dioxide. Phenol red is the pH indicator. Ammonia raises the pH, causing phenol red to turn from orange-red below pH 8.4 (Fig. 11.7 Tube A) to magenta pink at a pH greater than 8.4 (Fig. 11.7 Tube C). Bacteria that acidify the media below pH 6.2 cause phenol red to turn yellow (Fig. 11.7 Tube B).

IMViC Tests
Together, the indole, methyl red (MR), Voges-Proskauer (VP), and citrate tests comprise the IMViC tests. This panel of tests is important in distinguishing Enterobacteriaceae members from each other. Table 1 shows the commonly encountered species in the Enterobacteriaceae and the typical test results.
Table 11.1: Typical Enterobacteriaceae culture results.
| Genus | Indole | MR | VP | Citrate | H2S | Motility | Lactose fermentation | Gelatin hydrolysis |
| Salmonella | – | + | – | + | + | + | – | – |
| Proteus | v | + | – | – | + | + | – | + |
| Klebsiella | – | – | + | + | – | – | + | – |
| Enterobacter | – | – | + | + | – | + | + | – |
| Citrobacter | + | + | – | + | + | + | + | – |
| Escherichia | + | + | – | – | – | + | + | – |
| Serratia | – | – | + | + | – | + | + | + |
| + indicates a positive reaction – indicates a negative reaction V indicates a variable reaction for species within a genus |
||||||||
Once you have all the test results for your organism, you then need to figure out which species you have. The table above can be useful, as can the more extensive table and dichotomous key on FOL. The dichotomous key is simply a series of yes/no questions about your test results. As you work through the questions, you will eventually arrive at a species name. Once you have a name, you can search to see if this species fits with your other observations. Remember, you need to justify the species that you ultimately arrive at in your conclusions.
Unknown Identification Exercise
Materials
- Gram staining kit
- Slides
- Cover slips
- Inoculating needle and loop
- Unknown plate
- Oxidase reagent
- Hydrogen peroxide
- Barritt’s A and B, Nitrate A and B, Kovac’s
- TSIA, SIM, nitrate broth, decarboxylase, MR-VP, phenol red glucose, phenol red lactose, Simmon’s citrate, gelatin
- Filter paper
Method
Colony Morphology
- From the streak plate, make observations on colony morphology.
Cellular morphology - Make a wet mount of the bacteria and observe cell arrangement and motility under the microscope.
- You will be testing for motility and cell arrangement using other tests; however, it is always best to have multiple tests to justify your conclusions.
- Gram stain: Make a Gram stain to confirm your culture is Gram negative. Make observations on cell morphology and arrangement.
Oxidase Test
- Soak a small filter paper in oxidase reagent in the lid of a petri plate. Use a loop to pick up bacteria from an agar plate and rub onto the filter paper. Record the colour of the bacterial culture. If there was no colour change within 2 minutes of smearing on the bacteria, then the bacteria are negative for the oxidase test.
Catalase Test
- Using a toothpick, pick up a cells (these can be from the first streaking on the plate) and smear onto a slide. Drop hydrogen peroxide onto the cells and observe for immediate bubble formation.
-
- You may need to use a magnifying glass to observe tiny bubbles if the strain is a weak positive.
Media
- Inoculate the following media: TSIA, SIM, gelatin (stab into the butt of the tube), phenol red glucose, phenol red lactose, Simmon’s citrate slant, nitrate broth, decarboxylase broth (with oil on the top of the tube), MR-VP tubes.
-
- VP: incubate 48 hours at 37 C. Aliquot 2.5 ml of culture into a new test tube. Add approximately 12 drops Barritt’s A (a-naphthol) and approximately 4 drops Barritt’s B (potassium hydroxide). Incubate 30 minutes. A red colour indicates the presence of acetoin.
- MR: incubate for 48 hours at 37 C. To the remaining 2.5 ml in the MR-VP tube, add 5 drops of methyl red. Interpret test results immediately- red indicates a positive result, yellow indicates a negative result.
- Gelatin hydrolysis: incubate 48 hours at 37 C. Gelatin liquefies at 28 C, so before reading the test result, place the gelatin tube and a control tube (which was also incubated, uninoculated, at 37 C) in an ice bath for 15 minutes. The control tube should be solid (if it isn’t, keep incubating in the ice bath until it is). Observe whether the test tube is solid or liquid. If you incubate too long on ice, all tubes will appear negative.
- SIM: Incubate for 24 hours at 37 C. Observe the presence of hydrogen sulphide (black precipitate) and motility (turbidity migrating from the stab). Drop Kovac’s reagent onto the tube and observe indole production as the reagent turns red.
- After 24 hours: Perform and interpret TSIA, phenol red tubes, nitrate broth, indole as previously described.
- After 48 hours, interpret the decarboxylase broth.
- Positive and negative controls: These will be inoculated by your professor and will be available for you to compare your results to.
Interpretation Using Media
- Use your results to determine the genus of your unknown culture.
-
- Is your unknown a member of the Enterobacteriaceae?
- If yes, you can use Table 1 and the table posted on FOL to see which is the best fit.
- Use the dichotomous key to arrive at a species.
- Look your species up on Microbe Wiki or in Bergey’s Manual of Determinative Bacteriology to determine if this genus is a good fit
Interpretation using DNA sequence - Using the DNA sequence provided from sequencing your PCR product or from your professor, search the DNA sequence into the BLAST search tool. Paste your sequence into the box under ‘Enter Query Sequence’, then hit the blue BLAST button at the bottom of the page. Examine the top hits with the most sequence identity with your sequence to determine the probable species.

