15.11: Ribosomes and Protein Synthesis - The Mechanism of Protein Synthesis
- Page ID
- 13312
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Describe the process of translation
As with mRNA synthesis, protein synthesis can be divided into three phases: initiation, elongation, and termination.
Initiation of Translation
Protein synthesis begins with the formation of a pre-initiation complex. In E. coli, this complex involves the small 30S ribosome, the mRNA template, three initiation factors (IFs; IF-1, IF-2, and IF-3), and a special initiator tRNA, called fMet-tRNA. The initiator tRNA basepairs to the start codon AUG (or rarely, GUG) and is covalently linked to a formylated methionine called fMet. Methionine is one of the 21 amino acids used in protein synthesis; formylated methionine is a methione to which a formyl group (a one-carbon aldehyde) has been covalently attached at the amino nitrogen. Formylated methionine is inserted by fMet-tRNA at the beginning of every polypeptide chain synthesized by E. coli, and is usually clipped off after translation is complete. When an in-frame AUG is encountered during translation elongation, a non-formylated methionine is inserted by a regular Met-tRNA. In E. coli mRNA, a sequence upstream of the first AUG codon, called the Shine-Dalgarno sequence (AGGAGG), interacts with the rRNA molecules that compose the ribosome. This interaction anchors the 30S ribosomal subunit at the correct location on the mRNA template.
In eukaryotes, a pre-initiation complex forms when an initiation factor called eIF2 ( eukaryotic initiation factor 2) binds GTP, and the GTP-eIF2 recruits the eukaryotic initiator tRNA to the 40s small ribosomal subunit. The initiator tRNA, called Met-tRNAi, carries unmodified methionine in eukaryotes, not fMet, but it is distinct from other cellular Met-tRNAs in that it can bind eIFs and it can bind at the ribosome P site. The eukaryotic pre-initiation complex then recognizes the 7-methylguanosine cap at the 5′ end of a mRNA. Several other eIFs, specifically eIF1, eIF3, and eIF4, act as cap-binding proteins and assist the recruitment of the pre-initiation complex to the 5′ cap. Poly (A)-Binding Protein (PAB) binds both the poly (A) tail of the mRNA and the complex of proteins at the cap and also assists in the process. Once at the cap, the pre-initiation complex tracks along the mRNA in the 5′ to 3′ direction, searching for the AUG start codon. Many, but not all, eukaryotic mRNAs are translated from the first AUG sequence. The nucleotides around the AUG indicate whether it is the correct start codon.
Once the appropriate AUG is identified, eIF2 hydrolyzes GTP to GDP and powers the delivery of the tRNAi-Met to the start codon, where the tRNAi anticodon basepairs to the AUG codon. After this, eIF2-GDP is released from the complex, and eIF5-GTP binds. The 60S ribosomal subunit is recruited to the pre-initiation complex by eIF5-GTP, which hydrolyzes its GTP to GDP to power the assembly of the full ribosome at the translation start site with the Met-tRNAi positioned in the ribosome P site. The remaining eIFs dissociate from the ribosome and translation is ready to begins.
In archaea, translation initiation is similar to that seen in eukaryotes, except that the initiation factors involved are called aIFs (archaeal inititiaion factors), not eIFs.
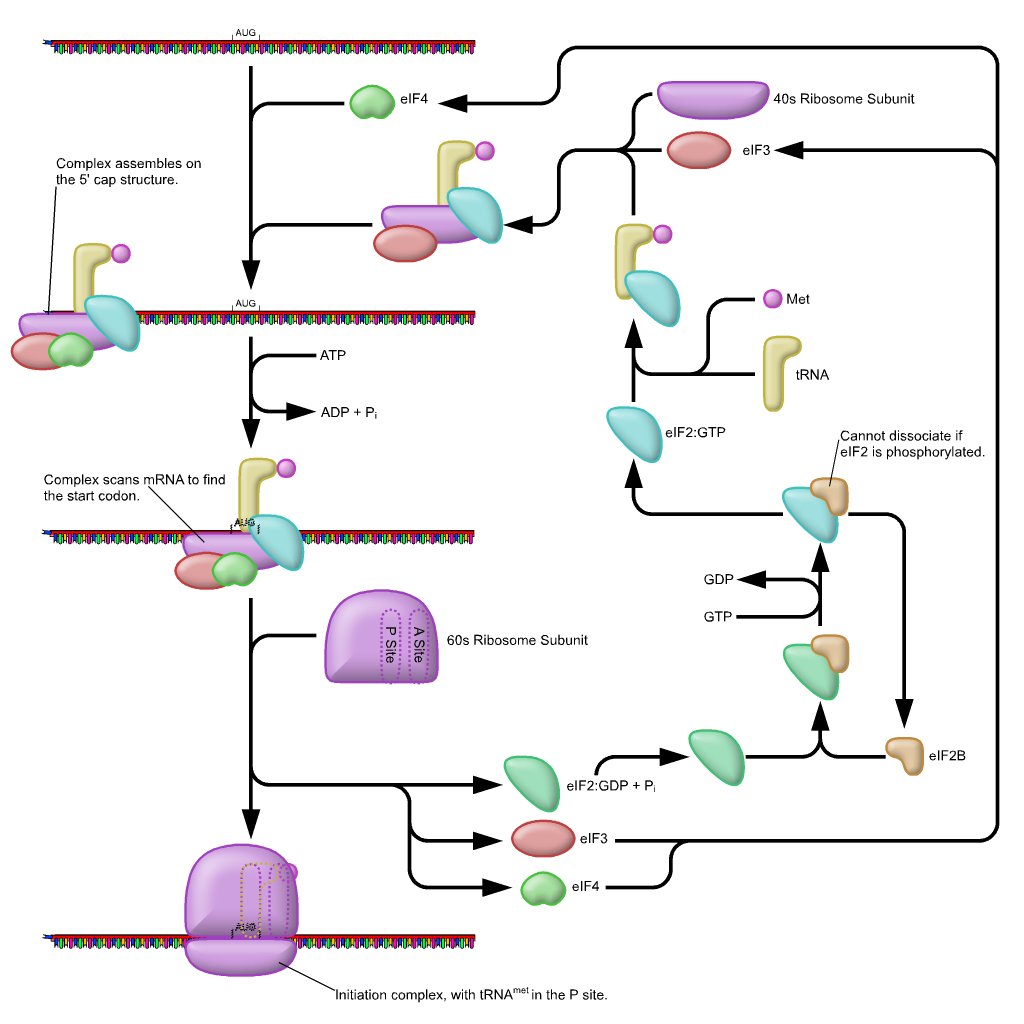
Translation Elongation
The basics of elongation are the same in prokaryotes and eukaryotes. The intact ribosome has three compartments: the A site binds incoming aminoacyl tRNAs; the P site binds tRNAs carrying the growing polypeptide chain; the E site releases dissociated tRNAs so that they can be recharged with amino acids. The initiator tRNA, rMet-tRNA in E. coli and Met-tRNAi in eukaryotes and archaea, binds directly to the P site. This creates an initiation complex with a free A site ready to accept the aminoacyl-tRNA corresponding to the first codon after the AUG.
The aminoacyl-tRNA with an anticodon complementary to the A site codon lands in the A site. A peptide bond is formed between the amino group of the A site amino acid and the carboxyl group of the most-recently attached amino acid in the growing polypeptide chain attached to the P-site tRNA.The formation of the peptide bond is catalyzed by peptidyl transferase, an RNA-based enzyme that is integrated into the large ribosomal subunit. The energy for the peptide bond formation is derived from GTP hydrolysis, which is catalyzed by a separate elongation factor.
Catalyzing the formation of a peptide bond removes the bond holding the growing polypeptide chain to the P-site tRNA. The growing polypeptide chain is transferred to the amino end of the incoming amino acid, and the A-site tRNA temporarily holds the growing polypeptide chain, while the P-site tRNA is now empty or uncharged.
The ribosome moves three nucleotides down the mRNA. The tRNAs are basepaired to a codon on the mRNA, so as the ribosome moves over the mRNA, the tRNAs stay in place while the ribosome moves and each tRNA is moved into the next tRNA binding site. The E site moves over the former P-site tRNA, now empty or uncharged, the P site moves over the former A-site tRNA, now carrying the growing polypeptide chain, and the A site moves over a new codon. In the E site, the uncharged tRNA detaches from its anticodon and is expelled. A new aminoacyl-tRNA with an anticodon complementary to the new A-site codon enters the ribosome at the A site and the elongation process repeats itself. The energy for each step of the ribosome is donated by an elongation factor that hydrolyzes GTP.
Translation termination
Termination of translation occurs when the ribosome moves over a stop codon (UAA, UAG, or UGA). There are no tRNAs with anticodons complementary to stop codons, so no tRNAs enter the A site. Instead, in both prokaryotes and eukaryotes, a protein called a release factor enters the A site. The release factors cause the ribosome peptidyl transferase to add a water molecule to the carboxyl end of the most recently added amino acid in the growing polypeptide chain attached to the P-site tRNA. This causes the polypeptide chain to detach from its tRNA, and the newly-made polypeptide is released. The small and large ribosomal subunits dissociate from the mRNA and from each other; they are recruited almost immediately into another translation initiation complex. After many ribosomes have completed translation, the mRNA is degraded so the nucleotides can be reused in another transcription reaction.
Modeling translation: This interactive models the process of translation in eukaryotes.
Key Points
- Protein synthesis, or translation, begins with a process known as pre-initiation, when the small ribosmal subunit, the mRNA template, initiator factors, and a special initiator tRNA, come together.
- During translocation and elongation, the ribosome moves one codon 3′ down the mRNA, brings in a charged tRNA to the A site, transfers the growing polypeptide chain from the P-site tRNA to the carboxyl group of the A-site amino acid, and ejects the uncharged tRNA at the E site.
- When a stop or nonsense codon (UAA, UAG, or UGA) is reached on the mRNA, the ribosome terminates translation.
Key Terms
- translation: a process occurring in the ribosome in which a strand of messenger RNA (mRNA) guides assembly of a sequence of amino acids to make a protein


