9.3: Signaling Molecules and Cellular Receptors - Types of Receptors
- Page ID
- 13216
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Compare internal receptors with cell-surface receptors
Types of Receptors
Receptors are protein molecules in the target cell or on its surface that bind ligands. There are two types of receptors: internal receptors and cell-surface receptors.
Internal receptors
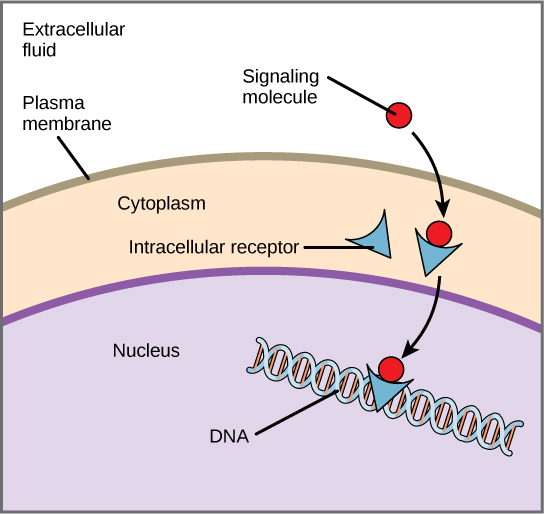
Internal receptors, also known as intracellular or cytoplasmic receptors, are found in the cytoplasm of the cell and respond to hydrophobic ligand molecules that are able to travel across the plasma membrane. Once inside the cell, many of these molecules bind to proteins that act as regulators of mRNA synthesis to mediate gene expression. Gene expression is the cellular process of transforming the information in a cell’s DNA into a sequence of amino acids that ultimately forms a protein. When the ligand binds to the internal receptor, a conformational change exposes a DNA-binding site on the protein. The ligand-receptor complex moves into the nucleus, binds to specific regulatory regions of the chromosomal DNA, and promotes the initiation of transcription. Internal receptors can directly influence gene expression without having to pass the signal on to other receptors or messengers.
Cell-Surface Receptors
Cell-surface receptors, also known as transmembrane receptors, are cell surface, membrane-anchored, or integral proteins that bind to external ligand molecules. This type of receptor spans the plasma membrane and performs signal transduction, converting an extracellular signal into an intracellular signal. Ligands that interact with cell-surface receptors do not have to enter the cell that they affect. Cell-surface receptors are also called cell-specific proteins or markers because they are specific to individual cell types.
Each cell-surface receptor has three main components: an external ligand-binding domain (extracellular domain), a hydrophobic membrane-spanning region, and an intracellular domain inside the cell. The size and extent of each of these domains vary widely, depending on the type of receptor.
Cell-surface receptors are involved in most of the signaling in multicellular organisms. There are three general categories of cell-surface receptors: ion channel-linked receptors, G-protein-linked receptors, and enzyme-linked receptors.
Ion Channel-Linked Receptors
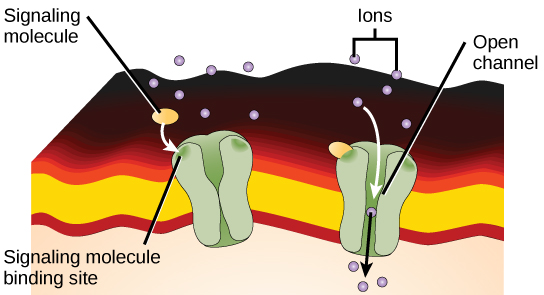
Ion channel-linked receptors bind a ligand and open a channel through the membrane that allows specific ions to pass through. To form a channel, this type of cell-surface receptor has an extensive membrane-spanning region. In order to interact with the phospholipid fatty acid tails that form the center of the plasma membrane, many of the amino acids in the membrane-spanning region are hydrophobic in nature. Conversely, the amino acids that line the inside of the channel are hydrophilic to allow for the passage of water or ions. When a ligand binds to the extracellular region of the channel, there is a conformational change in the protein’s structure that allows ions such as sodium, calcium, magnesium, and hydrogen to pass through.
G-Protein Linked Receptors
G-protein-linked receptors bind a ligand and activate a membrane protein called a G-protein. The activated G-protein then interacts with either an ion channel or an enzyme in the membrane. All G-protein-linked receptors have seven transmembrane domains, but each receptor has its own specific extracellular domain and G-protein-binding site.
Cell signaling using G-protein-linked receptors occurs as a cyclic series of events. Before the ligand binds, the inactive G-protein can bind to a newly-revealed site on the receptor specific for its binding. Once the G-protein binds to the receptor, the resultant shape change activates the G-protein, which releases GDP and picks up GTP. The subunits of the G-protein then split into the α subunit and the β subunit. One or both of these G-protein fragments may be able to activate other proteins as a result. Later, the GTP on the active α subunit of the G-protein is hydrolyzed to GDP and the β subunit is deactivated. The subunits reassociate to form the inactive G-protein, and the cycle starts over.

Enzyme-Linked Receptors
Enzyme-linked receptors are cell-surface receptors with intracellular domains that are associated with an enzyme. In some cases, the intracellular domain of the receptor itself is an enzyme or the enzyme-linked receptor has an intracellular domain that interacts directly with an enzyme. The enzyme-linked receptors normally have large extracellular and intracellular domains, but the membrane-spanning region consists of a single alpha-helical region of the peptide strand. When a ligand binds to the extracellular domain, a signal is transferred through the membrane and activates the enzyme, which sets off a chain of events within the cell that eventually leads to a response. An example of this type of enzyme-linked receptor is the tyrosine kinase receptor. The tyrosine kinase receptor transfers phosphate groups to tyrosine molecules. Signaling molecules bind to the extracellular domain of two nearby tyrosine kinase receptors, which then dimerize. Phosphates are then added to tyrosine residues on the intracellular domain of the receptors and can then transmit the signal to the next messenger within the cytoplasm.
Key Points
- Intracellular receptors are located in the cytoplasm of the cell and are activated by hydrophobic ligand molecules that can pass through the plasma membrane.
- Cell-surface receptors bind to an external ligand molecule and convert an extracellular signal into an intracellular signal.
- Three general categories of cell-surface receptors include: ion -channel, G- protein, and enzyme -linked protein receptors.
- Ion channel -linked receptors bind a ligand and open a channel through the membrane that allows specific ions to pass through.
- G-protein-linked receptors bind a ligand and activate a membrane protein called a G-protein, which then interacts with either an ion channel or an enzyme in the membrane.
- Enzyme-linked receptors are cell-surface receptors with intracellular domains that are associated with an enzyme.
Key Terms
- integral protein: a protein molecule (or assembly of proteins) that is permanently attached to the biological membrane
- transcription: the synthesis of RNA under the direction of DNA


