3.3: The Nucleus
- Page ID
- 3967
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The nucleus is the hallmark of eukaryotic cells; the very term eukaryotic means having a "true nucleus".
The Nuclear Envelope
The nucleus is enveloped by a pair of membranes enclosing a lumen that is continuous with that of the endoplasmic reticulum. The inner membrane is stabilized by a meshwork of intermediate filament proteins called lamins. The nuclear envelope is perforated by thousands of nuclear pore complexes (NPCs) that control the passage of molecules in and out of the nucleus.
Chromatin
 The nucleus contains the chromosomes of the cell. Each chromosome consists of a single molecule of DNA complexed with an equal mass of proteins. Collectively, the DNA of the nucleus with its associated proteins is called chromatin.
The nucleus contains the chromosomes of the cell. Each chromosome consists of a single molecule of DNA complexed with an equal mass of proteins. Collectively, the DNA of the nucleus with its associated proteins is called chromatin.
Most of the protein consists of multiple copies of 5 kinds of histones. These are basic proteins, bristling with positively charged arginine and lysine residues. (Both Arg and Lys have a free amino group on their R group, which attracts protons (H+) giving them a positive charge.) Just the choice of amino acids you would make to bind tightly to the negatively-charged phosphate groups of DNA.
Chromatin also contains small amounts of a wide variety of nonhistone proteins. Most of these are transcription factors (e.g., the steroid receptors) and their association with the DNA is more transient.
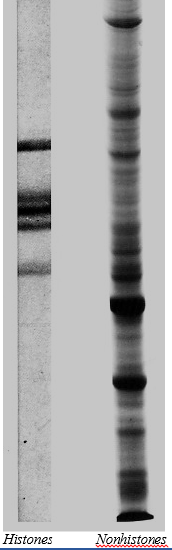
The image on the right shows the 5 histones separated by electrophoresis. These 5 proteins vary little from one cell type to another or even from one species to another. However, the many nonhistone proteins in chromatin (also shown on the right) do vary from one cell type to another and from one species to another. (Courtesy of Gary S. Stein and Janet Swinehart Stein, University of Florida.)
Two copies of each of four kinds of histones H2A, H2B, H3, H4 form a core of protein, the nucleosome core. Around this is wrapped about 147 base pairs of DNA. From 20–60 bp of DNA link one nucleosome to the next. Each linker region is occupied by a single molecule of histone 1 (H1). This region is longer (50–150 bp) adjacent to the promoters of genes which presumably makes more room for the binding of transcription factors.
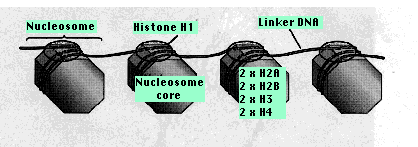
Nucleosomes Nucleosome Schematics
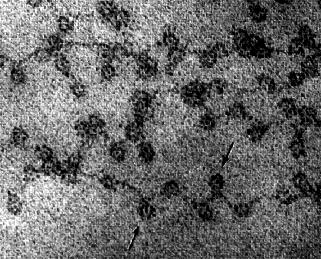
The binding of histones to DNA does not depend on particular nucleotide sequences in the DNA but does depend critically on the amino acid sequence of the histone. Histones are some of the most conserved molecules during the course of evolution. Histone H4 in the calf differs from H4 in the pea plant at only 2 amino acids residues in the chain of 102. The above electron micrograph (courtesy of David E. Olins and Ada L. Olins) for nucleosomes shows chromatin from the nucleus of a chicken red blood cell (birds, unlike most mammals, retain the nucleus in their mature red blood cells). The arrows point to the nucleosomes. You can see why the arrangement of nucleosomes has been likened to "beads on a string".
The formation of nucleosomes helps somewhat, but not nearly enough, to make the DNA sufficiently compact to fit in the nucleus. In order to fit 46 DNA molecules (in humans), totaling over 2 meters in length, into a nucleus that may be only 10 µm across requires more extensive folding and compaction. Interactions between the exposed "tails" of the core histones causes nucleosomes to associate into a compact fiber 30 nm in diameter. These fibers are then folded into more complex structures whose precise configuration is uncertain and which probably changes with the level of activity of the genes in the region.
Histone Modifications
Although their amino acid sequence (primary structure) is unvarying, individual histone molecules do vary in structure as a result of chemical modifications that occur later to individual amino acids. These include adding:
- acetyl groups (CH3CO−) to lysines
- phosphate groups to serines and threonines
- methyl groups to lysines and arginines
Although 75–80% of the histone molecule is incorporated in the core, the remainder — at the N-terminal — dangles out from the core as a "tail" (not shown in the figure). Most of the chemical modifications occur on these tails, especially of H3 and H4. Most of theses changes are reversible. For example, acetyl groups are added by enzymes called histone acetyltransferases (HATs)(not to be confused with the "HAT" medium used to make monoclonal antibodies) and are also removed by histone deacetylases (HDACs). More often than not, acetylation of histones occurs in regions of chromatin that become active in gene transcription. This makes a kind of intuitive sense as adding acetyl groups neutralizes the positive charges on Lys thus reducing the strength of the association between the highly-negative DNA and the highly-positive histones.
However, there is surely more to the story. Acetylation of Lys-16 on H4 ("H4K16ac") prevents the interaction of their "tails" needed to form the compact 30-nm structure of inactive chromatin and thus is associated with active genes (note that this case involves interrupting protein-protein not protein-DNA interactions). Methylation, which also neutralizes the charge on lysines (and arginines), can either stimulate or inhibit gene transcription in that region.
- Adding 3 methyl groups to lysine-4 and/or lysine-36 in H3 (H3K4me3 and H3K36me3 respectively) is associated with active gene transcription while
- trimethylation of lysine-9 and/or lysine-27 in H3 (H3K9me3 and H3K27me3 respectively) is associated with inactive genes. (These include those imprinted genes that have been permanently inactivated in somatic cells.)
- And adding phosphates causes the chromosomes to become more — not less — compact as they get ready for mitosis and meiosis.
In any case, it is now clear that histones are a dynamic component of chromatin and not simply inert DNA-packing material.
Histone Variants
We have genes for 8 different varieties of histone 1 (H1). Which variety is found at a particular linker depends on such factors as the type of cell, where it is in the cell cycle, and its stage of differentiation. In some cases, at least, a particular variant of H1 associates with certain transcription factors to bind to the enhancer of specific genes turning off expression of those genes.
Some other examples of histone variants:
- H3 is replaced by CENP-A ("centromere protein A") at the nucleosomes near centromeres. Failure to substitute CENP-A for H3 in this regions blocks centromere structure and function.
- H2A is replaced by the variant H2A.Z at gene promoters and enhancers.
- All the "standard" histones are replaced by variants as sperm develop.
In general, the "standard" histones are incorporated into the nucleosomes as new DNA is synthesized during S phase of the cell cycle. Later, some are replaced by variant histones as conditions in the cell dictate.
Chromosome Territories
During interphase, little can be seen of chromatin structure (except for special cases like the polytene chromosomes of Drosophila and some other flies). Although each chromosome is greatly elongated, it tends to occupy a discrete region within the nucleus called its territory. This can be demonstrated by:
- directing a tiny laser beam at a small portion of the nucleus. If all the chromosomes were intertwined, one would expect that all would receive some damage. That does not occur — only one or two chromosomes are damaged.
- Fluorescent stains specific for a particular chromosome stain only two regions in the nucleus — revealing the territory of the two homologs.
"Kissing" Chromosomes
Portions of one chromosome can loop out of its territory and interact with part of a different chromosome looping out from its territory. These are "kissing" chromosomes. The examples that have been found so far indicate that these interactions are another way of coordinating the activity of genes residing on different chromosomes.
The human genome contains many genes — scattered along different chromosomes — that are turned on by the arrival of a single signal. Among the many genes activated by estrogen, are TIFF1 on chromosome 21 and GREB1 on chromosome 2. Using FISH analysis, researchers at the University of California in San Diego showed that within a little as 2 minutes after exposing cells to estrogen, the TIFF1 and GREB1 loci move from their respective chromosome territories and "kiss".
In the mouse, naive helper T cells — awaiting a signal to direct them to become either Th1 cells or Th2 cells have
- the part of chromosome 10 carrying the gene for interferon-gamma (a Th1 cytokine) kissing
- the part of chromosome 11 carrying the genes for IL-4 and IL-5 (Th2 cytokines).
When the cell receives the signals committing it to one path or the other, the two regions separate, the appropriate one going to a region of active transcription; the other to a region of heterochromatin.
And still another example (in this case, two loci far apart on the same chromosome "kiss"):
In the head region of the Drosophila larva, expression of the homeobox (HOX) genes Antp and Abd-B is shut down. FISH analysis shows that these two loci — 10,000,000 base pairs apart on chromosome III — are brought together in the nucleus bound by proteins that prevent their transcription.
Euchromatin versus Heterochromatin
The density of the chromatin that makes up each chromosome (that is, how tightly it is packed) varies along the length of the chromosome. Dense regions are called heterochromatin and less dense regions are called euchromatin. Heterochromatin is found in parts of the chromosome where there are few or no genes, such as
- centromeres and
- telomeres. This heterochromatin is found in all types of cells in the organism.
- is also found in gene-rich regions of the chromosome but where the genes are inactive; that is, not transcribed. The location of this heterochromatin varies from one type of differentiated cell to another (as we would expect — a liver cell, for example, should shut down expression of genes that are not needed for its functions.
- is densely-packed.
- is greatly enriched with transposons and other types of DNA that does not contribute to the proteome.
- is replicated late in S phase of the cell cycle.
- has reduced crossing over in meiosis.
- is localized near the inner surface of the nuclear envelope, in most animal cells.
- The histones in the nucleosomes of heterochromatin show characteristic modifications:
- decreased acetylation;
- increased methylation of lysine-9 in histone H3 (H3K9), which now provides a binding site for heterochromatin protein 1 (HP1), which blocks access by the transcription factors needed for gene transcription
- increased methylation of lysine-27 in histone H3 (H3K27)
Euchromatin
- is found in parts of the chromosome that contain many active genes.
- is loosely-packed in loops of 30-nm fibers.
- separated from adjacent heterochromatin by insulators.
- In animal cells, euchromatin and thus active gene transcription occurs near the center of the nucleus.
- The genes in euchromatin show
- decreased methylation of the cytosines in CpG sites of the gene's promoter(s)
- increased acetylation of nearby histones
- decreased methylation of lysine-9 and lysine-27 in histone H3
Nucleosomes and Transcription
Transcription factors cannot bind to their promoter if the promoter is blocked by a nucleosome. One of the first functions of the assembling transcription factors is to either expel the nucleosome from the site where transcription begins or at least to slide the nucleosomes along the DNA molecule. Either action exposes the gene's promoter so that the transcription factors can then bind to it.
The actual transcription of protein-coding genes is done by RNA polymerase II (Pol II or RNAP II). In order for it to travel along the DNA to be transcribed, a complex of proteins removes the nucleosomes in front of it and then replaces them after Pol II has transcribed that portion of DNA and moved on.
Nucleosomes and DNA Replication
As is the case in transcription, the DNA helix must open to allow DNA replication to proceed. This, too, requires that the nucleosomes preceding the replication fork be removed and then quickly reassembled as the leading and lagging strands are synthesized.
The Nucleolus
During the period between cell divisions, when the chromosomes are in their extended state, one or more of them (10 in human cells) have loops extending into a spherical mass called the nucleolus. Here are synthesized three (of the four) kinds of RNA molecules (28S, 18S, 5.8S) used in the assembly of the large and small subunits of ribosomes.
28S, 18S, and 5.8S ribosomal RNA is transcribed (by RNA polymerase I) from hundreds to thousands of tandemly-arranged rDNA genes distributed (in humans) on 10 different chromosomes. The rDNA-containing regions of these 10 chromosomes cluster together in the nucleolus.
(In yeast, the 5S rRNA molecules — as well as transfer RNA molecules — are also synthesized (by RNA polymerase III) in the nucleolus.)
Once formed, rRNA molecules associate with the dozens of different ribosomal proteins used in the assembly of the large and small subunits of the ribosome.
But proteins are synthesized in the cytosol — and all the ribosomes are needed in the cytosol to do their work — so there must be a mechanism for the transport of these large structures in and out of the nucleus. This is one of the functions of the nuclear pore complexes.
Nuclear Pore Complexes (NPCs)
 The nuclear envelope is perforated with thousands of pores. Each is constructed from multiple copies of several dozen different proteins called nucleoporins.
The nuclear envelope is perforated with thousands of pores. Each is constructed from multiple copies of several dozen different proteins called nucleoporins.
The entire assembly forms an aqueous channel connecting the cytosol with the interior of the nucleus ("nucleoplasm"). When materials are to be transported through the pore, it opens up to form a channel some 27–41 nm wide — large enough to get such large assemblies as ribosomal subunits through.
Transport through the nuclear pore complexes is active; that is, it requires
- energy
- many different carrier molecules each specialized to transport a particular cargo
- docking molecules in the NPC (represented here as colored rods and disks)
Import into the nucleus
Proteins are synthesized in the cytosol and those needed by the nucleus must be imported into it through the NPCs. They include:
- all the histones needed to make the nucleosomes
- all the ribosomal proteins needed for the assembly of ribosomes
- all the transcription factors (e.g., the steroid receptors) needed to turn genes on (and off)
- all the splicing factors needed to process pre-mRNA into mature mRNA molecules; that is, to cut out intron regions and splice the exon regions.
Probably all of these proteins has a characteristic sequence of amino acids — called a nuclear localization sequence (NLS) — that target them for entry.
Export from the nucleus
Molecules and macromolecular assemblies exported from the nucleus include:
- the ribosomal subunits containing both rRNA and proteins
- messenger RNA (mRNA) molecules (accompanied by proteins)
- transfer RNA (tRNA) molecules (also accompanied by proteins)
- transcription factors that are returned to the cytosol to await reuse
Both the RNA and protein molecules contain a characteristic nuclear export sequence (NES) needed to ensure their association with the right carrier molecules to take them out to the cytosol.
Contributors and Attributions
John W. Kimball. This content is distributed under a Creative Commons Attribution 3.0 Unported (CC BY 3.0) license and made possible by funding from The Saylor Foundation.


