9: Evolvability and Plasticity
- Page ID
- 14439
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Evolvability has emerged as the connection between population-level evolution and macroevolution. Although both of these processes are fundamentally the same (both occurring through mutation, drift, and selection), they happen at such different scales that we typically use very different tools and mindsets to study each of them. For example, population-level evolution is often modeled with Hardy-Weinberg related equations examining the fitness and prevalence of particular alleles. On the other hand, we typically study macroevolutionary change by comparing distinct species using the fossil record and molecular genetics. The question is, then, how do we go from here to there? How do these micro-level processes eventually result in macro-level patterns? Evolvability is one useful way to approach this. A population is "evolvable" if it can cope with changing environments via adaptation. That is, if genetic mutations in the population have a good chance of increasing fitness.
While at the surface level, this seems simplistic, we can follow it to deeper levels to see how the evolvability of a population affects developmental processes whose evolution underlies macroevolutionary changes. Gerhart and Kirschner tackle this in their paper the Theory of Facilitated Variation. In this paper they identify major routes by which developmental genetic changes can "facilitate" (or help increase) phenotypic variation without compromising fitness (i.e. they increase evolvability).
These routes, termed weak regulatory linkage, exploratory processes, and compartmentation, are summarized below:
Ability to engage in weak regulatory linkage
Linkage occurs between processes that are connected to each other or to the same specific conditions. For example, when you are cold you shiver and get goosebumps. These two processes are connected to each other via a condition (cold) and also via the sympathetic nervous system. Likewise, developmental processes can be linked via shared gene regulatory networks and/or via shared regulatory components (for example genes that are expressed in response to the same transcription factor, or signal transduction pathways that respond to the same signaling molecule).
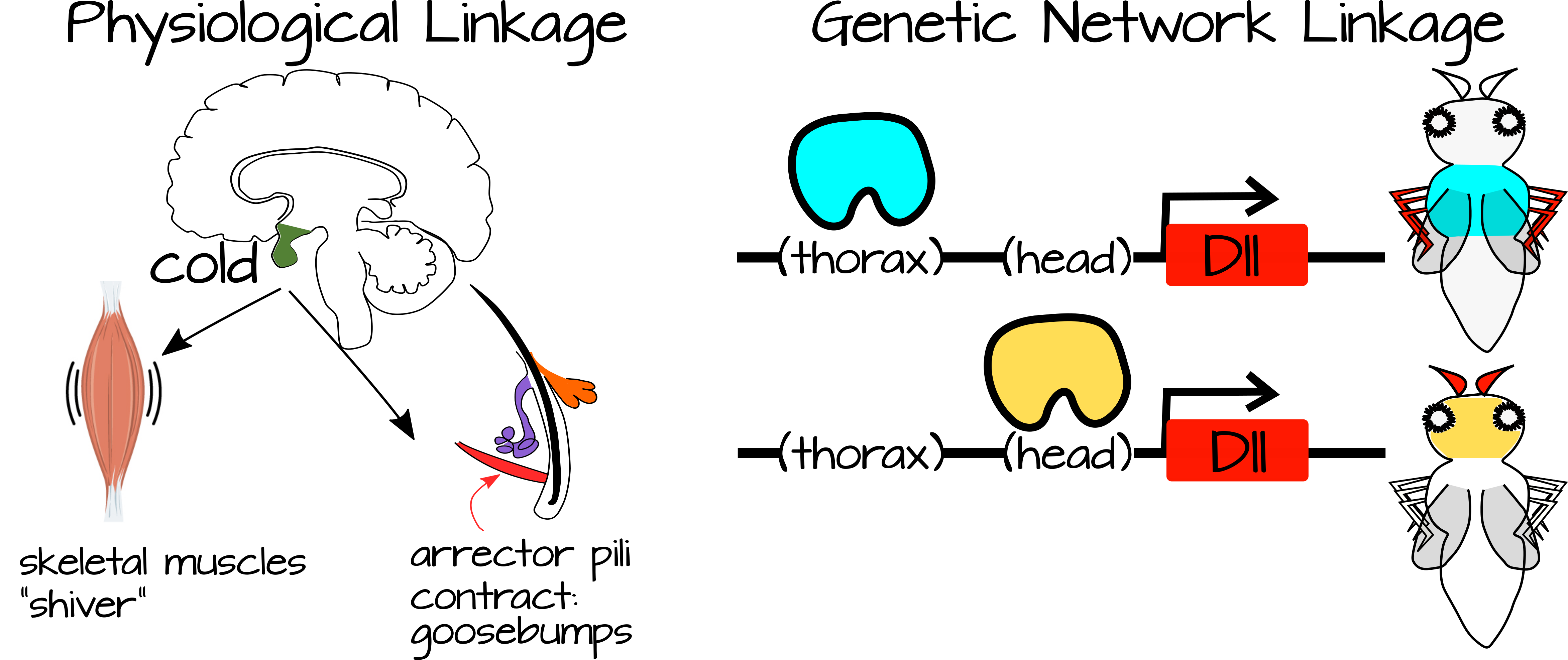
Weak regulatory linkage refers to linked processes that are regulated by simple inputs that do not provide much information to the processes. We have seen examples of this in Habits of Highly Effective Signalling Pathways, where a transcription factor releases repression on a set of downstream genes. These genes self-activate via local activators as soon as the inhibition is released suggesting there could potentially be multiple ways to de-inhibit them. This is considered weak regulatory linkage because the regulation by the transcription factor is "weak." When local activators are present, the default for the process is "on". The transcription factor is only needed to release inhibition. Once this inhibition is released, a cascade of gene expression can occur, resulting in a complex process with large developmental or physiological consequences. In this way a complex process can be turned off or turned on in new places with small-scale regulatory changes. For example, the transcription factor expression can be turned up or down, or other proteins can act to modify the ability of the transcription factor to bind to its target DNA.
In general, weak regulatory linkage occurs when a simple signal can trigger multiple complex processes depending on the cellular context. Gerhart and Kirschner note that this increases the plasticity of a system since small changes to the regulatory factors (transcription factors for example) can change the functional output of a complex system. The complex system itself is largely self-regulating, requiring only a trigger from the regulatory factors. In this way, development can occur more slowly or more quickly in certain conditions, choices between two tissue states (e.g. gonad type) can be modified by environmental factors, etc.
Exploratory processes
Exploratory processes are search and find processes like following a chemical cue gradient to a point source. We see this type of process in development when we examine how axons form during neurogenesis and how vasculature arises to feed organs and tissues. We see it in adults in the vertebrate adaptive immune system and foraging behavior in chemically-guided organisms like ants. We say that this type of process shows robustness because it adapts quickly to local environmental changes. For example, with strict XY grid patterning of vasculature, growth over developmental time would need to be under strict control and size/shape variance in organs would not be tolerated. Small blood vessels, however, do not grow due to a coordinate system, rather they grow based on a "supply and demand" system with outgrowths to regions of low oxygen. Cells secrete a protein signal (VEGF) when they are low on oxygen, promoting blood vessel growth towards them2.
Exploratory processes are also adaptable over evolutionary time since they allow for size and shape variance. This is important within populations, where individuals may vary in size or shape. But it is also important over longer periods of evolutionary time as these flexible patterning processes will form a useable system based on their simple set of rules for growth. For example, transplanted leg discs in insects become innervated as do ectopic limbs in chick embryos3,4. In this way, exploratory processes facilitate evolution by helping to build a viable body when other morphological components have evolved into more fit conformations.
Compartmentation
In plants, the ability to separate the the light reactions from the Calvin Cycle of photosynthesis either temporally (as in CAM plants) or spatially (as in C4 plants) has lead to incredible success and diversity in these plant types as they are no longer reliant on keeping their stomata open during the hottest and dryest (but also sunniest) parts of the day. In our own cells, we can see compartmentalization of our mitochondria, with the intermembrane space about 10 times more acidic than the lumen. This allows for the proton motive force to be concentrated and also protects the pH of the rest of the cell. Our cells also physically separate cell-cell signaling from transcriptional regulation - allowing modification of signaling pathways dependent on cell history (development) and environment.
When one compartment of an organism (or cell) can act semi-independently of another, we expect higher variation. That is, each compartment can run its developmental program via activation and inhibition of specific GRNs without disrupting the development of other compartments. In this way, gene expression changes that affect only one or a few compartments might be limited in effect. Evolution thus may be able to act on compartments individually, as long as these are expression changes like transcript number, protein isoforms, or post-translational modification. Protein-coding mutations, on the other hand, could potentially act on many compartments at once. Evolution can potentially act on individual compartments via gene expression changes, while exploratory processes can maintain the robustness of signaling and nutrient pathways across the entire body.
Evolvability in animals relies on two competing types of development:
- Processes resistant to evolutionary change. These include exploratory processes that use external signals to find the most efficient or most effective morphology, leading to retention of the process of. But these can also include processes that are conserved over long evolutionary periods of time due to physical or genetic constraints.
- Changable processes. These include processes that change over time because they are either compartmentalized or are co-opted onto other robust developmental programs. Compartmentation (a process resistant to change) allows for physical separation of developmental processes, and weak regulatory linkage allows for co-option of processes to new times and places (for example compartments).
In this way, evolutionary change happens on a background of robust processes. Some of these robust processes are malleable in developmental time (like vascularization), while others are constraints - limiting what is possible in a particular evolutionary lineage. One example we have already considered is tetrapod limb evolution. While there are physical and genetic constraints on this limiting the number of bone condensations per developmental section, these constraints also create compartments - the stylopod, zeugopod, and autopod as well as the 5 digits. Size variation and adaptations to the compartments is supported by robust exploratory processes, linking bone development to supporting musculature, vasculature, and innervation.
Further Reading
Deep homology (genetic constraints): https://www.sdbonline.org/sites/fly/lewhelddeep/deephomology.htm
Bauplans (developmental constraints): https://evolution-outreach.springeropen.com/articles/10.1007/s12052-012-0424-z
Genetics is modulated by environment, this modulation can be short-lived (like getting a tan) or long-lived (like getting insulin-resistant diabetes). It can even span generations (thrifty epigenotype). How does this "plasticity" arise in genetics and how does this change how evolution works?
http://science.sciencemag.org/content/357/6350/eaan0221.long: If you get your 23andme genotype done, it will only tell you what your risk is for certain disease and disorders. Why can't we be sure?
References
- "The Theory of Facilitated Variation" 2007, PNAS, John Gerhart and Marc Kirschner, https://doi.org/10.1073/pnas.0701035104
- Reviewed in "Hypoxia-Induced Angiogenesis: Good and Evil," 2011, Genes and Cancer, Bryan L. Krock, Nicolas Skuli. and M. Celeste Simon doi: 10.1177/1947601911423654
- "Axonal projections from transplanted ectopic legs in an insect," 1985, Journal of Comparative Neurology, P. Sivasubramanian D. R. Nässel, doi.org/10.1002/cne.902390211
- "The innervation of FGF-induced additional limbs in the chick embryo," 2003, Journal of Anatomy, BW Turney, AM Rowan-Hull, and JM Brown doi: 10.1046/j.1469-7580.2003.00131.x
- "Angiogenesis: An Adaptive Dynamic Biological Patterning Problem," 2013, PLoS Computational Biology, Timothy W. Secomb , Jonathan P. Alberding, Richard Hsu, Mark W. Dewhirst, Axel R. Pries https://doi.org/10.1371/journal.pcbi.1002983
- "Neuronal migration and lamination in the vertebrate retina," 2018, Frontiers in Neuroscience, Rana Amini, Mauricio Rocha-Martins, and Caren Norden. doi: 10.3389/fnins.2017.00742
Thumbnail is a graph of standing variation in a population (10,000 polymorphic sites from Human Chromosome 1) by Graham Coop, published on Wikimedia Commons under a CC BY 3.0 license.


