15: In situ hybridization
( \newcommand{\kernel}{\mathrm{null}\,}\)
Summary
In situ hybridization shows where in a tissue sample an RNA of a specific sequence is found or where in a set of chromosomes a DNA of a specific sequence is found.
Also known as
In situ, ISH
Usually when a specific DNA sequence (instead of RNA) is the target, this method is referred to as fluorescence in situ hybridization, or FISH. Usually, if someone refers to the method as just “in situ,” they mean they are probing for RNA, and if they say “FISH,” they mean they’re probing for DNA, but it’s best to clarify.
Samples needed
Fixed whole tissue, fixed tissue sections, or fixed cells
Method
Here, we will focus on how in situ is used to probe for an RNA sequence in a tissue sample, but the principles are the same across applications. First, the sample is chemically fixed, meaning that any living cells are killed and all metabolic activity ceases, and the tissue structure is chemically preserved. Next, the sample is permeabilized to allow the probe access to the nucleic acids in the sample. The sample is then incubated with a nucleic acid probe complementary to the RNA of interest. The probe binds the RNA transcript if present. The probe must be attached to another molecule that allows for visualization, whether that be a radioactive label, fluorescent label, etc.
Controls
The probe binding specificity should be tested through the use of positive and negative controls.
Interpretation
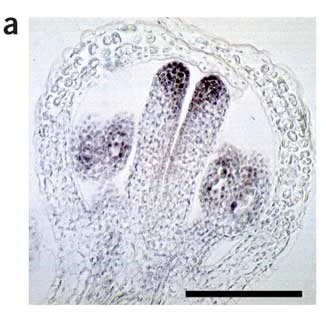
Figure 1. In situ hybridization for SPATULA mRNA. Relevant section of caption for published figure reads: “(a) Localization of SPATULA mRNA in a developing Arabidopsis flower (longitudinal section). Note the expression in the tips of the two carpels. Signal can also be seen in the anthers.” “Figure 1” by Brewer et al.[1]. [Image description]
This image comes from a methods paper detailing how to perform in situ hybridization on Arabidopsis tissues, but a very similar photograph was generated in another study by some of the same researchers on SPATULA function[2]. The study showed that SPATULA encodes a transcription factor that controls the growth of certain reproductive structures during flower development, including the carpels, which produce female gametes, and anthers, which produce male gametes. One step in showing that SPATULA regulates the growth of these structures was demonstrating that SPATULA is expressed there. The in situ hybridization results in Figure 1 display staining in the relevant structures, indicating that SPATULA mRNA is present.
Image Descriptions
Figure 1 image description:
A stained longitudinal section of a developing flower. The tissue is shaped like a hollow bulb with two finger-like projections inside, and one more bulbous projection on either side of the fingers. The “fingers” are the carpels, and strong staining can be seen at the “fingertips.” The bulbs are anthers, which have lighter staining.
Thumbnail
"Xenopus laevis embyro.jpg"↗ by Emily is licensed under CC BY 4.0↗.
Description: In situ hybridization of MRF4 (myf6) gene in X. laevis embryo.
Author
Katherine Mattaini, Tufts University
-
Brewer, P. B., M. G. Heis ler, J. Hejátko, J. Friml, and E. Benková. 2006. In situ hybridization for mRNA detection in Arabidopsis tissue sections. Nature Protocols 1:1462–1467. ↵
-
Heisler, M. G. B., A. Atkinson, Y. H. Bylstra, R. Walsh, and D. R. Smyth. 2001. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128:1089–1098. ↵

