7: Column Chromatography
- Page ID
- 141652
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Summary
Column chromatography is a method for separating a mixture of substances based on their chemical and physical characteristics. In biological research, this is usually a mixture of proteins.
Also known as
Variants: Affinity chromatography, ion exchange chromatography, size exclusion chromatography
Samples needed
In biology, most column chromatography is performed on a solution containing a mixture of proteins
Method
Although column chromatography is used widely for many applications in chemistry, in biology and biochemistry, it is most often used to separate an aqueous mixture of proteins. The column is a tube filled with a stationary phase, also known as a resin. A solution called the mobile phase is then run through the column, either by gravity, or under pressure. The separation of the mixture into component proteins is based on the difference in how each protein interacts with the stationary phase vs. the mobile phase. For instance, a protein that interacts strongly with the stationary phase will take longer to move from the beginning to the end of the column, and subsequently, it will be eluted from the column later than another protein that interacts only weakly with the stationary phase. The liquid phase that comes off the end of the column can be collected in a series of fractions, which can then be used for further applications.
There are many variants of column chromatography that differ by the characteristic of the protein that is used to separate the mixture.
Ion exchange chromatography
Ion exchange chromatography separates proteins based on their overall charge. In this method, the stationary phase is either positively or negatively charged. The mobile phase is a solution that has a low salt concentration initially, but over the course of the run, the concentration of salt in the mobile phase is increased so that proteins that were initially strongly attracted to the stationary phase are eluted. For instance, if the stationary phase is negatively charged, proteins that have an overall negative charge will run through the column very quickly. However, proteins with an overall positive charge will interact with the stationary phase and therefore have a longer elution time. As the salt concentration increases, proteins that are weakly positively charged will elute first, followed by those that are strongly positively charged.
Size exclusion chromatography
As the name implies, size exclusion chromatography separates proteins based on their size and shape, and it is also called gel filtration. The resin in size exclusion chromatography is made of beads that have pores or channels running through them. The size of the beads and the channels can be used to fine-tune the separation achieved by this method. Large proteins will not be able to fit into the channels in the beads, and will therefore have the shortest elution times. However, smaller proteins will get caught up in the winding channels, and therefore, they will have a longer path through the column and longer elution times.
The outcome of size exclusion chromatography is the opposite of gel electrophoresis, which can be confusing! In size exclusion chromatography, large proteins move the fastest through the column, whereas in gel electrophoresis, they move through the gel the slowest, since the gel acts as a simple sieve.
Affinity chromatography
Affinity chromatography achieves separation based on the affinity of one or a small number of proteins for a particular substance affixed to the stationary phase. For instance, if ATP is affixed to the stationary phase, ATP-binding proteins would interact with it, and non-ATP-binding proteins would be quickly eluted. The ATP-binding proteins could be eluted by changing the mobile phase to a buffer containing “free” (not resin-bound) ATP. Another extremely common example is purification of proteins with a 6xHis epitope tag with a Ni2+ or Co2+ column. This method is known as immobilized metal affinity chromatography, or IMAC. Some of the N atoms in the histidine ring coordinate the metal ion, so that tagged proteins are bound to the column, and untagged proteins flow through. The tagged proteins can be eluted using a buffer containing imidazole, which has the same chemical structure as the ring in the amino acid histidine. Finally, if a column is derivatized with an antibody that binds a specific protein, that can also be considered affinity chromatography. However, when purifying a protein using an antibody, immunoprecipitation is often performed instead of chromatography.
Interpretation
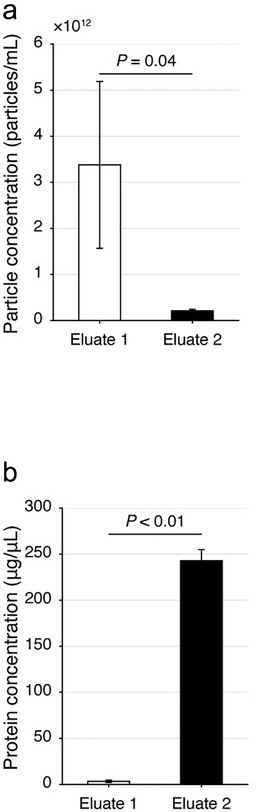 Figure 1. Analysis of extracellular vesicles (EVs) and proteins from human serum in eluate 1, collected early in a size exclusion chromatography experiment, vs. in eluate 2, collected later in the experiment. Relevant section of caption for published figure reads: “Evaluation of the dichotomic SEC method using human serum. (a, b) The particle (a) and protein (b) concentrations of the Eluate 1 and 2 with the dichotomic SEC method. Data are shown with mean±SD, donor n = 3, Student's t-test.” “Figure 1” by Gul et al.[1] [Image description]
Figure 1. Analysis of extracellular vesicles (EVs) and proteins from human serum in eluate 1, collected early in a size exclusion chromatography experiment, vs. in eluate 2, collected later in the experiment. Relevant section of caption for published figure reads: “Evaluation of the dichotomic SEC method using human serum. (a, b) The particle (a) and protein (b) concentrations of the Eluate 1 and 2 with the dichotomic SEC method. Data are shown with mean±SD, donor n = 3, Student's t-test.” “Figure 1” by Gul et al.[1] [Image description]In this experiment, researchers showed that they successfully developed a relatively simple protocol to separate extracellular vesicles (EVs) from soluble proteins in human serum. Obviously, extracellular vesicles are far larger than individual proteins, so the use of size exclusion chromatography was a good choice. To make the protocol as straightforward as possible, all the mobile phase eluted from the column was divided into only two “fractions,” eluate 1 and eluate 2. Through extensive protocol optimization, the researchers were able to establish a cutoff between eluates 1 and 2 that resulted in good separation between the EVs and the proteins from the serum. In panel a, they measured the amount of particles, i.e. EVs, in each eluate using a special instrument. The instrument determined particle concentration through a mixture of light scattering and Brownian motion. In panel b, they measured protein concentration of each eluate using a Bradford assay. As expected, EVs were far more abundant in eluate 1, since they are larger and would not get caught up in the channels of the resin. Eluate 2 contained most of the proteins not associated with the EVs, since they were much smaller.
Image Descriptions
Figure 1 image description:
Two column graphs. Graph a shows particle concentration. Eluate 1 contains ~3.4 x 1012 ± 0.8 x 1012 particles/ml and Eluate 2 contains ~0.25 x 1012 ± 0.05 x 1012 particles/ml. p = 0.04. Graph b shows protein concentration in μg/μl. Eluate 1 contains ~5 ± 1 μg/μl and Eluate 2 contains ~240 ± 20 μg/μl. p < 0.01. ↵
Thumbnail
"Chromatography column.PNG"↗ by Klaas1978 is in the Public Domain↗.
Description: Chromatography column with sample in three different stages.
Author
Katherine Mattaini, Tufts University
-
Guo, J., C. Wu, X. Lin, J. Zhou, J. Zhang, W. Zheng, T. Wang, and Y. Cui. 2021. Establishment of a simplified dichotomic size-exclusion chromatography for isolating extracellular vesicles toward clinical applications. Journal of Extracellular Vesicles 10:e12145. ↵

