10.1: Introduction
- Page ID
- 24885
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)History of Genetic Transformation
Any uptake of genetic information from the external environment into cells that results in the expression of new traits is called genetic transformation. This process can occur naturally. Some bacteria are referred to as being “competent” to indicate that they are capable of taking DNA into the cell from the environment. This is referred to as natural competence. Bacteria are also capable of receiving DNA through the process of conjugation where plasmids from one bacteria are sent to another through the conjugation pilus. Other methods of introduction of foreign DNA include direct injection into the cytosol or through the use of viruses in a process called transduction. In eukaryotic cells, we refer to the introduction of DNA as transfection.
Frederick Griffith and the Transforming Agent
At the beginning of modern biology, the source of genetic material was not known to be DNA. In fact, many scientists thought DNA was too simple to perform this job. Scientists believed that proteins, with their 20 varied amino acids, were the carriers of genetic information. In an attempt to develop a vaccine for bacterial-induced pneumonia, Frederick Griffith was the first to describe the process of genetic transformation by accident in 1928. Griffith took a virulent strain of bacteria (smooth in appearance) that caused pneumonia and injected them into mice. This would result in the death of the mice. He also observed that injection of a rough bacteria did not cause any disease. After heat-killing the smooth bacteria, he discovered that living bacteria of the virulent strain was required for the disease to progress. Finally, he observed that injecting the heat-killed virulent bacteria with living bacteria of the non-virulent strain resulted in pneumonia and death in the mice. From this experiment, a transforming agent with the capacity to pass on a trait was found to be within the contents of those dead cells. But no one knew this agent to be DNA at that point.

Hershey and Chase
Hershey and Chase studied bacteriophage (phage=eater). Phages are bacterial viruses that infect bacteria and cause lysis of the cells. They have a very simple structure of a proteinaceous head/collar/tail and a DNA core. It was known that bacteria infected with phage were resistant to additional infection. In 1952 Hershey and Chase grew bacteriophage in conditions that would specifically label either the DNA or the protein with radioactivity. They subsequently took phage with radiolabeled DNA and infected bacteria. In parallel, they took phage with radiolabeled protein and infected another set of bacteria. After just enough time for infection, the bacterial cultures were placed into a blender to separate the bacteriophage from the bacteria.
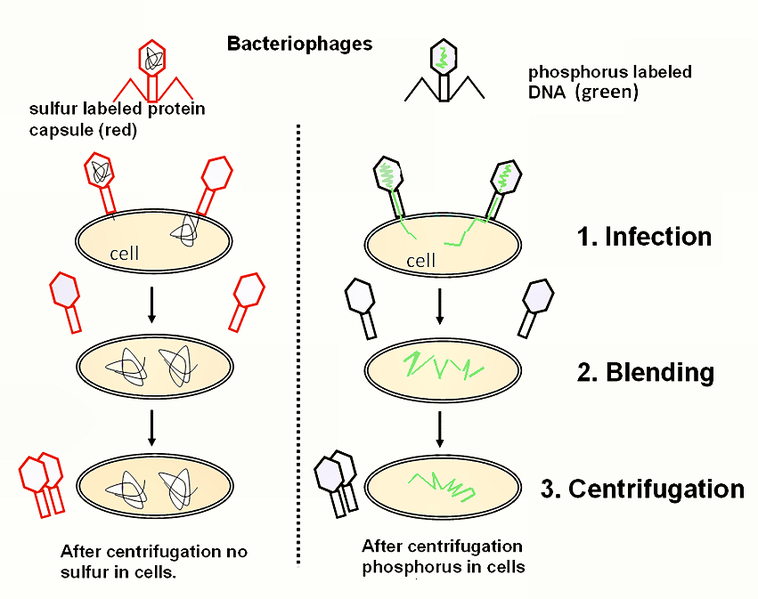
Solutions were centrifuged to isolate bacteria from the phage. Bacteria were radioactive only when the phage grown in conditions to radiolabel DNA infected the bacteria to indicate that DNA might be the transforming agent.
Questions to Think About
- What isotope would be used to label protein?
- What isotope would be used to label just DNA?
Avery, McCarty, and MacCleod
In 1944, Oswald Avery, Colin MacCleod and Maclyn McCarty repeated Griffith’s experiment. Instead of using heat-killed bacteria, these scientists isolated protein, carbohydrates, lipids and nucleic acids from the virulent strain and co-injected with the non-virulent bacteria. Carbohydrate extracts were ineffective at transforming bacteria. Protein extracts were incapable of causing transformation. Lipid injections were unable to result in virulence. Only nucleic acid samples treated with RNase were capable of transforming bacteria. When co-injecting with DNase, bacteria were not transformed. Along with Hershey-Chase, this definitively illustrated that DNA was the transforming agent capable of transferring genetic information.



