3.7: Enzyme Kinetics with Spectrovis
- Page ID
- 24753
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Enzyme Kinetics of Turnip Peroxidase
Hydrogen peroxide (H2O2) is a strong oxidizing agent that can damage cells and is formed as a by-product of oxygen consumption. Fortunately, aerobic cells contain peroxidases that break down peroxide into water and oxygen. This enzyme reduces hydrogen peroxide into H2O by oxidizing an organic compound (AH2 -> A).
This exercise uses turnip extract as a source of peroxidase. This turnip extract requires a source of electrons (a reducing agent) in order for the reaction to occur. In this case, a colorless organic compound called guaiacol is used. Guaiacol is oxidized in the process of converting the peroxide and becomes brown. The enzymatic activity can then be traced using a spectrophotometer to measure the amount of brown being formed.
Set-up and Calibrate LabQuest with SpectroVis Plus
- Set-up the cuvette to serve as a Blank.
- Add 10 drops 0.02% hydrogen peroxide.
- Add 5 drops 0.2% guaiacol.
- Add 20 drops of pH 7 buffer.
- Add 10 drops extraction buffered.
- Connect a LabQuest2 to a SpectroVis plus and start the data collection program.
- Calibrate the SpectroVis.
- Change Mode.
- Select “Time-Based” from the dropdown menu and press OK when done.
- Change Mode.
- Rate: 0.5 samples/s
- Interval: 2 s/sample
- Duration: 200 s
2. Press the Play button (green arrow).
1. Perform Warm-up for 90 seconds.
1. Choose "Finish Calibration".
2. Press "OK".
2. Press the "Stop" button (Red Square).
4. Select “Meter Icon” (top left most of the display).
- Press the large red area showing the current absorbance reading.
- Choose “Change Wavelength”.
- Set Wavelength to 500nm.
Effect of pH on Peroxidase Activity
- Set-up the cuvette for pH 3.
- Add 10 drops 0.02% hydrogen peroxide.
- Add 5 drops 0.2% guaiacol.
- Add 20 drops of pH 3 buffer.
- Add 10 drops turnip extraction last.
- Quickly invert the cuvette and place the cuvette into the Spectrovis Plus.
- Press play button to begin recording data (choose “discard data” if prompted).
- After 200s, remove SpectroVis from USB.
- Insert a USB flash drive and wait 1 minute.
- Press File → Export → choose USB icon and rename the file and add “.csv” to the end of the file name.
- Press OK.
- Sequentially repeat the experiment exchanging the pH buffer with pH 5, 7, 10.
Effect of Temperature on Peroxidase Activity
- Set-up the cuvette for 0ºC
- Add 10 drops 0.02% hydrogen peroxide (incubated at 0ºC for 10 minutes).
- Add 5 drops 0.2% guaiacol.
- Add 20 drops of extraction buffer at 0ºC.
- Add 10 drops turnip extraction (incubated at 0ºC for 10 minutes) last.
- Quickly invert the cuvette and place the cuvette into the Spectrovis Plus.
- Press play button to begin recording data (choose “discard data” if prompted).
- After 200s, remove SpectroVis from USB.
- Insert a USB flash drive and wait 1 minute.
- Press File → Export → choose USB icon and rename the file and add “.csv” to the end of the file name.
- Press OK.
- Sequentially repeat the experiment exchanging the buffers stored at 20ºC, 40ºC, 60ºC.
Effect of Substrate Concentration on Peroxidase Activity
- Set-up the cuvette for 1X substrate.
- Add 10 drops 0.02% hydrogen peroxide.
- Add 5 drops 0.2% guaiacol.
- Add 20 drops of extraction buffer.
- Add 10 drops turnip extraction last.
- Quickly invert the cuvette and place the cuvette into the Spectrovis Plus.
- Press play button to begin recording data (choose “discard data” if prompted).
- After 200s, remove SpectroVis from USB.
- Insert a USB flash drive and wait 1 minute.
- Press File → Export → choose USB icon and rename the file and add “.csv” to the end of the file name.
- Press OK.
- Sequentially repeat the experiment with different amounts of buffer and peroxide:
- Repeat the experiment with 0.2X substrate and export data to a USB drive.
- Repeat the experiment with 2X substrate and export data to a USB drive.
- Repeat the experiment with 3X substrate and export data to a USB drive.

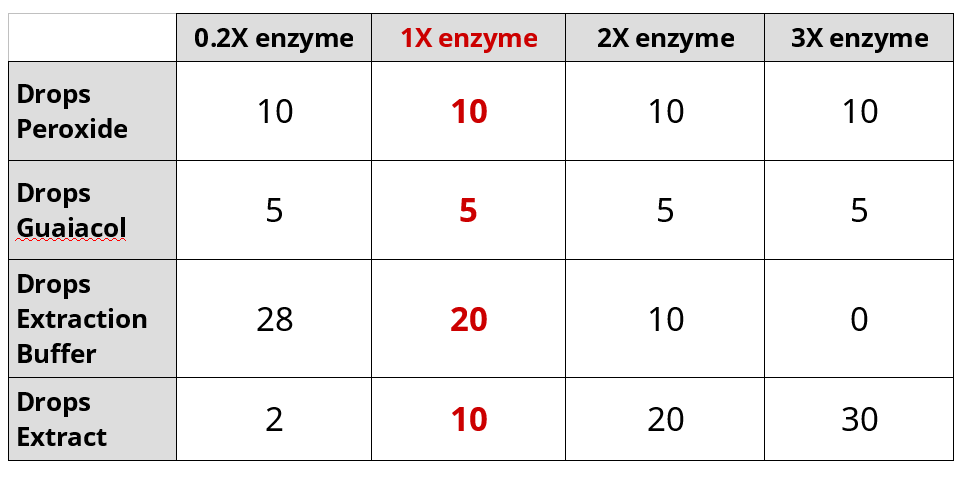
Effect of Enzyme Concentration on Peroxidase Activity
- Set-up the cuvette for 1X enzyme.
- Add 10 drops 0.02% hydrogen peroxide.
- Add 5 drops 0.2% guaiacol.
- Add 20 drops of extraction buffer.
- Add 10 drops turnip extraction last.
- Quickly invert the cuvette and place the cuvette into the Spectrovis Plus.
- Press play button to begin recording data (choose “discard data” if prompted).
- After 200s, remove SpectroVis from USB.
- Insert a USB flash drive and wait 1 minute.
- Press File → Export → choose USB icon and rename the file and add “.csv” to the end of the file name.
- Press OK.
- Sequentially repeat the experiment with different amounts of buffer and extract:
- Perform the experiment on 1X enzyme and export data to a USB drive.
- Repeat the experiment with 0.2X enzyme and export data to a USB drive.
- Repeat the experiment with 2X enzyme and export data to a USB drive.
- Repeat the experiment with 3X enzyme and export data to a USB drive.