2.6: The lac Operon; CAP site; DNA footprinting
- Page ID
- 18131
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The E. coli lac operon
.png?revision=1&size=bestfit&width=623&height=458)
Figure 2.6.1: lac Operon
- lac Z codes for b-galactosidase, which is an enzyme that cleaves b-galactosides (e.g. lactose).
- lac Y codes for permease, which is involved in the transport of b-galactosides into the cell.
- lac A codes for b-galactoside transacetylase, which acetylates b-galactosides.
- A mutation in either lac Z or lac Y can lead to a lac- genotype, i.e. cells which cannot utilize b-galactosides as a nutrient.
- A lac A- mutant, lacking transacetylase activity, can still utilize b-galactosides (it is still lac+ genotype). Its role in the metabolism of bgalactosides is not clear.
Promoter
a region of DNA involved in binding of RNA polymerase to initiate transcription.
Terminator
a sequence of DNA that causes RNA polymerase to terminate transcription.
- The cluster of three genes, lac ZYA, is transcribed into a single mRNA (polycistronic message) from a promoter just upstream from the lac Z gene.
- In the absence of an inducer the gene cluster is not transcribed.
- When an inducer is added (e.g. lactose, or the non-hydrolyzable analog isopropyl thiogalactoside - IPTG) transcription starts at a single promoter (lac P) and proceeds through the lac ZYA genes to a terminator sequence located downstream of the lac A gene.
Note
The lac ZYA mRNA has a half life of ~3 minutes, which allows induction to be reversed relatively rapidly (i.e. cells stop producing enzymes rapidly after induction stops).
Exercise 2.6.1
What molecule does the inducer (lactose) interact with to affect transcriptional regulation (i.e. induction of the lac operon)?
- Answer
-
It is not b-galactosidase, permase or transacetylase, rather it is a separate protein called a repressor protein.
- The lac genes are controlled by a mechanism called negative regulation.
- This means that they are transcribed unless they are turned off by the regulator protein.
- A mutation that inactivates the regulator protein causes the lacZYA genes to be continually expressed.
There are two types of genes in the lac operon:
- Structural genes - they code for enzymes required for some biochemical pathway (e.g. lac Z, Y and A).
- Regulator genes - they code for proteins involved in regulation of structural genes.
lac I is the regulator gene of the lac operon.
- This gene is located just upstream of the promoter region for the lac structural genes.
- The lac I gene has its own promoter (constitutive) and terminator.
- It makes a monocistronic message, and codes for one protein - the lac repressor protein.
- It can prevent transcription
- It can recognize and bind the small molecule inducer (lactose or IPTG)
Prevention of transcription by the lac repressor
- lac repressor (active as a tetrameric protein) binds to a sequence of DNA called the operator (lac O region).
- The operator region lies between the lac promoter region (site of RNA polymerase binding and transcription initiation) and the lac Z gene.
- The first 26 base pairs of the lac Z gene comprise the operator region.
- It is not that the repressor protein "blocks" the movement of RNA polymerase through the lac Z gene.
- Repressor binding and RNA polymerase binding (to the promoter) are mutually exclusive at the lac promoter/operator (lac PO) region.
How does the repressor/operator interaction change in the presence of the inducer molecule?
- The inducer can bind to the repressor to form a repressor/inducer complex that no longer associates with the operator.
- The key feature of this interaction is that the repressor protein has two binding sites, one for the inducer and one for the operator.
- When the inducer binds at its site, it changes the conformation of the repressor protein such that the operator binding site has a much reduced affinity for the DNA operator region.
- This type of control is called allosteric control.
- The result is that when the inducer is added the repressor is converted to a form which releases from the operator.
.png?revision=1&size=bestfit&width=709&height=436)
Figure 2.6.2: Inducer
Positive control of the lac operon is exerted by cAMP-CAP complex
- E. coli prefers glucose over other carbon sources.
- When E. coli is grown on glucose, if another sugar (e.g. lactose) is added the induction of enzymes to utilize the other sugar does not occur until the glucose is used up.
- When E. coli is starved for glucose it synthesizes an unusual nucleotide: cyclic 3'5' adenosine monophosphate (cyclic AMP, or cAMP):
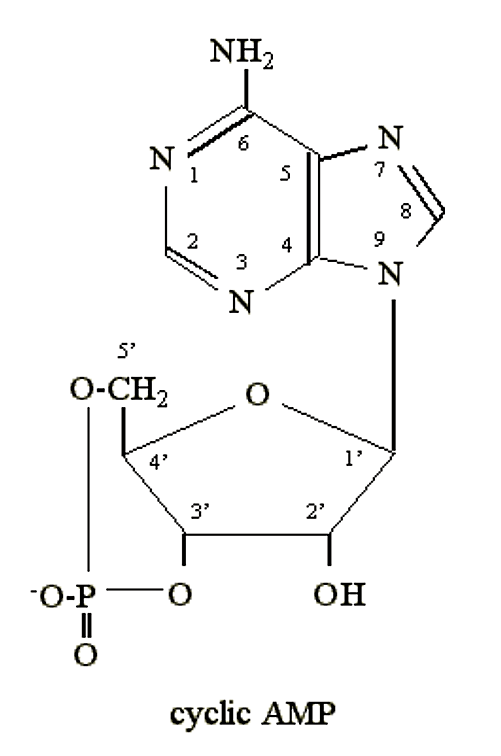.png?revision=1&size=bestfit&width=228&height=344)
Figure 2.6.3: cAMP
- In bacteria an increase in the cAMP level seems to be an "alert" signal indicating a low glucose level:
.png?revision=1&size=bestfit&width=527&height=219)
Figure 2.6.4: Interaction of cAMP level and lac operon
Dibutyryl cAMP
- an analogue of cAMP which can pass through the E. coli membrane and into the cell.
- If this is added to media containing glucose and lactose it will result in the induction of the lac operon.
- Thus, it mimics the chemical message which tricks the E. coli to respond as though glucose levels were low.
- Mutants of E. coli have been isolated which cannot be induced to metabolize any sugar other than glucose. There were two general categories of mutants:
- Class I. Defective in the enzyme adenylate cyclase. These mutants are unable to make cAMP even when the glucose conentration is low.
- Class II. Lacks a particular protein known as cAMP receptor protein (CRP) or, also known as catabolite receptor protein (CRP).
- Maximum transcription from the lac operon requires the presence of a cAMP/CRP complex.
- cAMP/CRP complex binds to a specific sequence in the lac control region called the "CAP" site.
- The CAP site is just upstream from the RNA polymerase binding site.
- Mutations in the CAP site that prevent cAMP-CRP binding also prevent high levels of expression of the lac operon.
- Thus, bound cAMP/CRP complex activates transcription (positive control), whereas bound lac repressor inhibits transcription (negative control).
- cAMP/CRP complex has affinity for DNA, and RNA pol.
- Enhances complex formation of RNA pol with the DNA promoter region.
Induction of the lac operon with lactose analogues
- The lac operon can be induced with lactose
- b-galactosidase (lacZ gene product) metabolizes the lactose
- When levels of lactose are reduced, the lac operon is again repressed by the lac repressor (lacI gene product)
- Non-metabolized lactose analogues can continually induce (i.e. de-repress) the lac operon
- isopropyl b-thiogalactoside, or IPTG, is a non-metabolized lactose analogue
DNA "footprinting" experiments
- If a protein binds to a region of DNA, it can protect that region of DNA from digestion by dnase (DNAse I: an endonuclease at sites adjacent to pyrimidine nucleotides).
- A fragment of DNA can be labeled at the 5' ends with 32P and then the label can be preferentially removed from one end (i.e. the 3' end of a gene) by an appropriate restriction endonuclease.
- If this DNA fragment, with a label at one specific end, forms a complex with a DNA binding protein the protein will protect the region of DNA that it binds to from DNAse I digestion.
- The digestion is done so as to be incomplete, for the purposes of this discussion, imagine that each DNA molecule is cleaved only once. Furthermore, the site of cleavage is randomly chosen from the available sites.
- Fragments of the DNA, separated and analyzed by size (using gel electrophoresis) after digestion will indicate the protected region:
.png?revision=1&size=bestfit&width=491&height=642)
Figure 2.6.5: DNA footprinting
Results of footprinting experiments
lac DNA incubated with either cAMP/CAP protein, or RNA polymerase, or lac I repressor protein:
.png?revision=1&size=bestfit&width=533&height=268)
Figure 2.6.6: lac repressor with cAMP
RNA polymerase interacts with specific promoter sequences and produces a "footprint" over a region of ~70 base pairs.
- This protection was observed to be more obvious on one strand than the other (i.e. if the other strand was labeled the results did not show as much protection).
- This region of DNAse protection included sites in the DNA from which mutagenesis experiments produced either "up" regulation or "down" regulation of promoter strength.
- These mutagenic "hot" spots affecting promoter strength were located at positions either -10 or -30 upstream from the transcription start site (position +1 in the above diagram):
.png?revision=1&size=bestfit&width=503&height=329)
Figure 2.6.7: Promoter Strength Mutations
- Promoters can be classified according to their "strength".
- This refers to the relative frequency of transcription initiation (transcriptional initiation events per minute), and is related to the affinity of RNA polymerase for the promoter region.
- Many promoters in E. coli have been characterized and a "consensus" promoter sequence has been identified:
.png?revision=2&size=bestfit&width=616&height=362)
Figure 2.6.8: Consensus Promoter Strength
Note
The lac promoter is a relatively weak promoter
RNA Polymerase
- E. coli RNA polymerase is a holoenzyme comprised of subunits b', b, a (dimer) and s70.
- The s70 subunit is the subunit which binds to the promoter region, but is unable to initiate RNA synthesis.
- After the s70 subunit subunit binds, the other subunits bind forming a function RNA polymerase.
- After approximately 10 base pairs have been transcribed the s70 subunit leaves and the core polymerase continues on.
The lac Operator Region
- The lac operator region is comprised of an (imperfect) inverted repeat region.
- Not surprisingly active repressor molecules are composed of a homodimer.
- In the homodimer structure there are a pair of a-helix regions which insert into adjacent major grooves of the DNA.
- The separation is approximately 34 angstroms apart.


