1.2: Structure of DNA and RNA
- Page ID
- 18120
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The Double Helix
DNA (deoxyribonucleic acid) and RNA (ribonucleic acid) are composed of two different classes of nitrogen-containing bases: the purines and pyrimidines. The most commonly occurring purines in DNA are adenine and guanine:
.png?revision=1&size=bestfit&width=542&height=439)
Figure 1.2.1: Purines
The most commonly occurring pyrimidines in DNA are cytosine and thymine:
.png?revision=1&size=bestfit&width=545&height=423)
Figure 1.2.2: Pyramidines
RNA contains the same bases as DNA with the exception of thymine. Instead, RNA contains the pyrimidine uracil:
.png?revision=1&size=bestfit&width=531&height=320)
Figure 1.2.3: Thymine vs. Uracil
Adenine, guanine, cytosine, thymine and uracil are usually abreviated using the single letter codes A, G, C, T and U, respectively.
Purines and pyrimidines can form chemical linkages with pentose (5-carbon) sugars. The carbon atoms on the sugars are designated 1', 2', 3', 4' and 5'. It is the 1' carbon of the sugar that becomes bonded to the nitrogen atom at position N1 of a pyrimidine or N9 of a purine. DNA precursors contain the pentose deoxyribose. RNA precursors contain the pentose ribose (which contains an additional OH group at the 2' position):
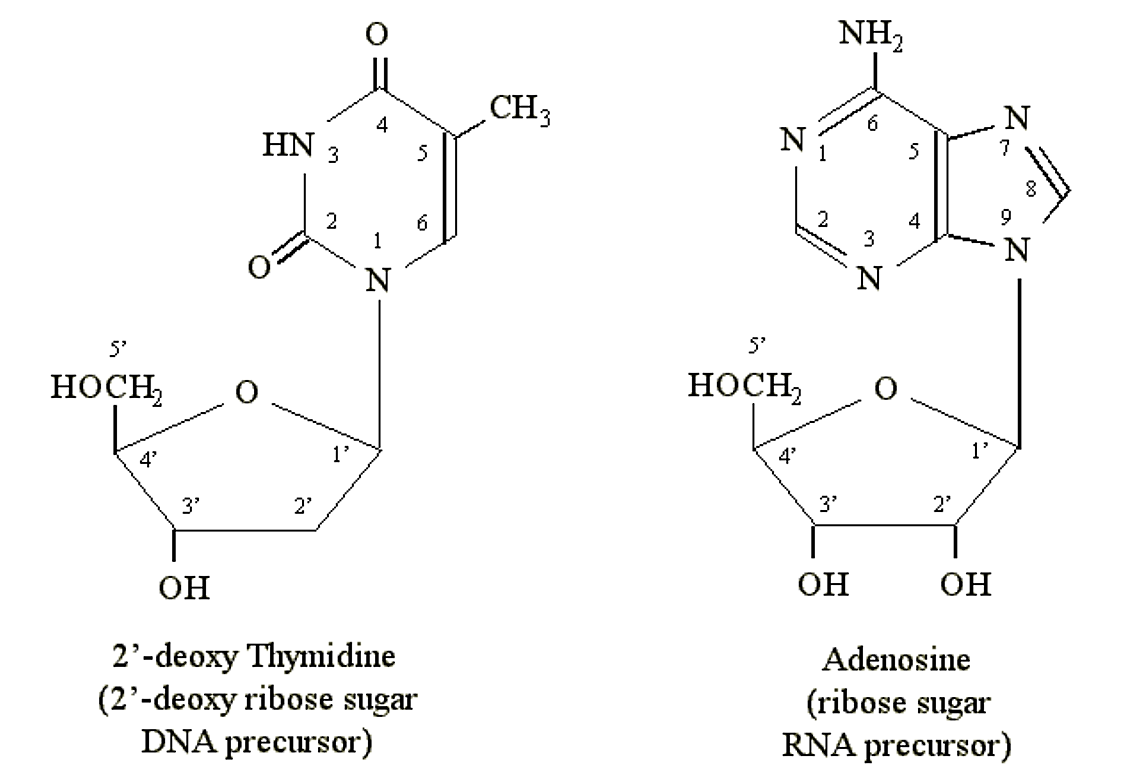.png?revision=1&size=bestfit&width=627&height=436)
Figure 1.2.4: Nucleosides
Before a nucleoside can become part of a DNA or RNA molecule it must become complexed with a phosphate group to form a nucleotide (either a deoxyribonucleotide or ribonucleotide). Nucleotides can posess 1, 2 or 3 phosphate groups, e.g. the nucleotides adenosine monophosphate (AMP), adenoside diphosphate (ADP) and adenosine triphosphate (ATP). The phosphate groups are attached to the 5' carbon of the ribose sugar moiety. Beginning with the phosphate group attached to the 5' ribose carbon, they are labeled a, b and g phosphate. It is the tri-phosphate nucleotide which is incorporated into DNA or RNA.
.png?revision=1&size=bestfit&width=407&height=353)
Figure 1.2.5: Nucleotide
DNA and RNA are simply long polymers of nucleotides called polynucleotides. Only the a phosphate is included in the polymer. It becomes chemically bonded to the 3' carbon of the sugar moiety of another nucleotide:
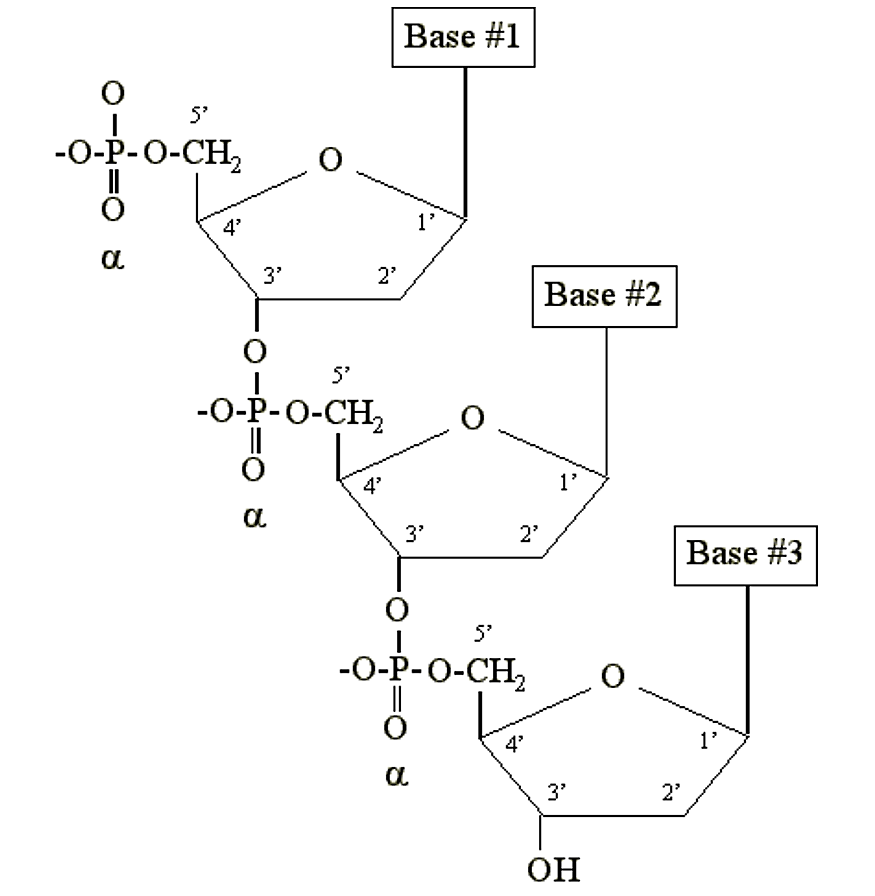.png?revision=2&size=bestfit&width=437&height=448)
.png?revision=1&size=bestfit&width=534&height=131)
Summary of terms:
|
|
|
|
|
|
|
|---|---|---|---|---|---|
| Adenine | Adenosine | (Adenylic acid) |
|
|
|
| Guanine | Guanosine | (Guanylic acid) |
|
|
|
| Cytosine | Cytidine | (Cytidylic acid) |
|
|
|
| Thymine | Thymidine | (Thymidylic acid) |
|
|
|
| Uracil | Uridine | (Uridylic acid) |
|
|
What is the structure of DNA? How is the structure related to function?
1950's
The primary chemical structure of polynucleotides was known (i.e. the 3'-5' phosphate linkage).
1951 E. Chargaff
The experiment:
Take DNA from a variety of species and hydrolyze it to yield individual pyrimidines and purines. Determine the relative concentrations of the A, T, C and G bases.
Result:
Although different species had uniquely different ratios of pyrimidines or purines, the relative concentrations of adenine always equaled that of thymine, and guanine equaled cytosine.
1950's R.E. Franklin
X-ray diffraction studies of DNA fibers demonstrated that DNA adopted a highly ordered helical structure. Franklin concluded that two or more chains must coil around each other to form a helix. Some basic dimensions of the helix were calculated from the x-ray diffraction data.
1953 L. Pauling and R.B. Corey
Propose a three chain helical structure for DNA with the phosphate backbone in the center and the bases on the outside.
1953 J.D. Watson and F.H.C. Crick
Identified a hydrogen bonding arrangement between models of thymine and adenine bases, and between cytosine and guanine bases which fullfilled Chargaff's rule:
.png?revision=1&size=bestfit&width=451&height=455)
Figure 1.2.7: Chargaff's Rule Bonding
Note that the "TA" pair can overlay the "GC" pair with the bonds to the sugar groups in similar juxtaposition. In the "double helix" model of Watson and Crick the polynucleotide chains interact to form a double helix with the chains running in opposite directions. The bases are directed towards the center (and stack on top of one another) and the sugar backbones face the outside of the helix.
The Watson and Crick model had the following physical dimensions:
- 34 Å per helical repeat
- 10 base pairs per repeat (i.e. per turn of the helix)
- 3.4 Å inter-base stacking distance
- 20 Å diameter for the helical width
Physical characteristics of the model matched those determined by Rosalind Franklin's x-ray diffraction studies.
Consequenses of the model for genetic information:
The Watson and Crick paper was an exercise in brevity (1 page only in Nature). The structure was so rich with implication that quite a bit could be written. The authors, however, chose only to say "It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material".
- If G always paired with C, and T always paired with A, then either strand could be regenerated from the complementary information in the other strand.
- The basis of the complementarity was hydrogen bonding, i.e. non-covalent interactions which could be easily broken and re-formed.
- The information which DNA carried was within the unique base sequence of the DNA.
- From the general interior location of the bases, it would appear that the double helix would have to dissociate in order to access the information.
- The non-equitorial location of the sugar moieties (see above) suggested that the DNA helix would have a major groove and a minor groove.
General notation of double stranded DNA:
.png?revision=1&size=bestfit&width=441&height=108)


