25.3: RNA-Dependent Synthesis of RNA and DNA
- Page ID
- 15199
RNA Viruses
Infections with RNA viruses place a constant burden on our healthcare systems and economy. Over the past century, this has been particularly true for infections with the Human immunodeficiency virus 1 (HIV-1), Influenza A virus (IAV), Rotavirus (RotaV), West Nile virus (WNV), Dengue virus (DV), Measles virus (MV), and the Porcine reproductive and respiratory syndrome virus (PRRSV). But also emergent RNA viruses can have considerable consequences, such as the Severe acute respiratory syndrome-related coronavirus (SARS-CoV) in 2003, the Middle East Respiratory Syndrome coronavirus (MERS-CoV), and most recently the Severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) in December of 2019.
Eukaryotes and bacteria can be infected with a wide variety of RNA viruses. On average, these pathogens share little sequence similarity and use different replication and transcription strategies. RNA virus genomes can consist of double-stranded RNA (dsRNA) or single-stranded (ssRNA) as shown in Figure \(\PageIndex{1}\) (Panel a). In turn, the ssRNA viruses can be classified into positive sense (+RNA) and negative sense (−RNA) viruses, depending on the translatability of their genetic material. As summarized for four model RNA pathogens in Panel b, all RNA viruses use dedicated replication and transcription strategies to amplify their genetic material. The common denominator of these strategies is a conserved RNA-dependent polymerase domain. Figure \(\PageIndex{1}\) describes variations of the replication process in the different types of RNA viruses.
Panel a shows a simplified taxonomy of the genome architecture of representative RNA viruses.
Panel b (+RNA virus)shows infection with a +RNA virus—as exemplified here with a CoV-like virion—releases a single-stranded RNA genome into the cytoplasm (1). (2) Translation of the 5′-terminal open-reading frame of the genome produces the viral replicase. (3) This multi-enzyme complex includes RNA-dependent RNA polymerase (RdRp) activity (orange) and associates with intracellular membranes before −RNA synthesis commences. Newly synthesized −RNAs are subsequently used to produce new +RNAs (4), which are typically capped (yellow) and polyadenylated (polyA). (Retrovirus) HIV-1 genomes are packaged as ssRNA in virions. When the ssRNA is released (1) a cDNA copy is synthesized by the reverse transcriptase enzyme (RT) (2). The RNA is next degraded by the intrinsic RNase H activity in the RT (3) and the single-stranded cDNA is converted to dsDNA (4). The dsDNA is imported into the nucleus (5) for integration into the host’s genetic material. (−RNA virus) (1) As illustrated here with an IAV-like particle, infection with an −RNA virus releases a viral RNA genome that is associated with a viral polymerase (orange) and nucleoprotein (green). (2) In the case of non-segmented −RNA viruses, these complexes support transcription to produce viral mRNAs or cRNAs. (3) Viral mRNAs are next translated and new viral proteins complex with cRNAs to synthesize new vRNAs. (5) The vRNA-containing complexes of some segmented −RNA viruses are imported into the nucleus of the host cell, where (6) the RdRp produces mRNAs or cRNAs. (7) mRNAs are transported to the cytoplasm, while cRNAs are bound by new viral proteins to form cRNPs for −RNA synthesis. (dsRNA virus) Fully duplexed RNA genomes lack cap and polyA elements. (1) The RdRp (orange), therefore, transcribes the viral genome inside the capsid of the virion (blue and red), so viral mRNAs can be (2) released into the cytoplasm as illustrated here with a rotavirus-like virion. In the cytoplasm, the mRNA is translated (3) or replicated by newly synthesized viral RdRps.
Viral RNA-dependent RNA Polymerases (RdRps) were discovered in the early 1960s from studies on mengovirus and polio viruses when it was observed that these viruses were not sensitive to actinomycin D, a drug that inhibits cellular DNA-directed RNA synthesis. This lack of sensitivity suggested that there is a virus-specific enzyme that could copy RNA from an RNA template and not from a DNA template. RdRps show some structural similarity to telomerase enzymes suggesting a potential ancestral relationship of telomerase with RdRps.
The most famous example of RdRp is of the polio virus. The viral genome is composed of RNA, which enters the cell through receptor-mediated endocytosis. From there, the RNA can act as a template for complementary RNA synthesis, immediately. The complementary strand is then, itself, able to act as a template for the production of new viral genomes that are further packaged and released from the cell ready to infect more host cells. The advantage of this method of replication is that there is no DNA stage; replication is quick and easy. The disadvantage is that there is no 'backup' DNA copy (Fig 26.3.1b; + RNA virus).
Reverse Transcriptase (RT) enzymes, on the other hand, that are common to retroviruses, convert the + RNA strand from the virus into a cDNA copy (Fig 26.3.1b; retroviruses). The RT enzyme then degrades the RNA and the single-stranded cDNA is converted to dsDNA. The dsDNA is then integrated into the host's genome where it can remain in a dormant state. Upon reactivation, + RNA will be manufactured, along with viral proteins used in the assembly of the infectious viral particles.
Structure and Mechanism of RNA Viral Polymerases
The RNA-dependent polymerase domain is a member of the superfamily of template-dependent nucleic acid polymerases and typically <400 amino acids in length. Its sequence is highly variable on average, with some regions showing less than ~10% conservation. Strong amino acid conservation can be observed, however, in regions that are directly involved in nucleotide selection or catalysis, such as the strictly conserved glycine and aspartates in the center of the domain. The prototypic RNA virus RNA polymerase domain harbors seven of such regions or motifs, which are arranged in the order G, F1–3, A, B, C, D, and E from amino- to carboxy-terminus. Each of the seven motifs in the RNA polymerase domain adopts a specific and conserved fold, as shown in Figure \(\PageIndex{2}\).
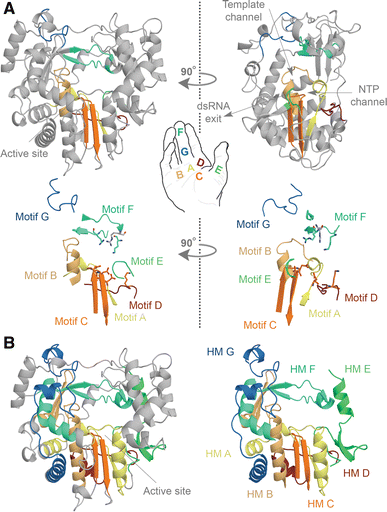
Panel a shows the structure of the FMDV RdRp. The motifs A, B, C, D, E, F, and G are color-coded yellow, gold, orange, red, light green, aquamarine, and blue. Overall the polymerase structure adopts a shape that resembles a cupped right hand. Herein, motifs A–E lie on the palm, while motifs F and G are part of the fingers. In the side-view of the enzyme, the location of the template and NTP channels is indicated.
Panel b shows conserved structural elements of the FMDV RdRp. Homomorphic A–G were mapped and color-coded yellow, gold, orange, red, light green, aquamarine, and blue, respectively. Images A and B are based on PDB accession 2E9R.
Figure \(\PageIndex{3}\) shows an interactive iCn3D model of the Foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA, PPi, and UTP (2E9Z)
Active site region side chains are shown as sticks and labeled. The template stand is shown in yellow and the primer is in green. Note the conserved amino acids Tyr-336, the catalytic Asp-338, and Lys-387 in motifs C and E, respectively. The primer 3'OH forms an H bond with active site Asp 338 (on motif C). In some structures, Asp 338 is bound to a metal ion that interacts with the PPP of the incoming NTP. Two metal ions are involved in catalysis. Tyr 336 interacts with the primer nucleotide while Lys 387 and Arg 388 interact with the primer backbone. These three residues are highly conserved. The acceptor base of the template strands is adjacent to the NTP binding site. There is no proofreading activity in viral RNA replication.
RNA-dependent RNA polymerases generally have a groove on top of the enzyme where RNA enters. It exits in the front. Nucleotides enter at a rear channel. This is illustrated in Figure \(\PageIndex{4}\) (Panel b). Following the convention for cellular DNA-dependent DNA polymerases (DdDp), the seven motifs and homomorphs are grouped into three subdomains. These subdomains are called fingers, palm, and thumb in reference to the polymerase domain’s likeliness to a cupped right hand, as shown in Figure \(\PageIndex{4}\) (Panel A). The finger subdomain loops that interconnect the fingers with the thumb in the RNA-dependent RNA polymerases (RdRps) of +RNA and dsRNA viruses create an overall “closed-hand” conformation that is unique to RNA-dependent RNA polymerases (RdRps) and generally not seen in crystal structures of DNA-dependent DNA polymerases (DdDps) or reverse transcriptases (RTs).
The three subdomains of the RNA-dependent polymerases work together to facilitate the binding of RNA and nucleotides (NTPs). The thumb subdomain contains various residues that are involved in RNA binding and these generally pack into the minor groove of the template RNA. In some polymerases, the thumb also contains sequences that protrude into the template channel to help stabilize the initiating NTPs on the ssRNA template. Crucially, these protrusions are also able to undergo relatively large conformational rearrangements to facilitate the translocation of the template following the first condensation reaction. The other residues of the thumb subdomain contribute to the formation of the NTP tunnel, which sits at an ~110° angle with the template channel as shown in Figure \(\PageIndex{4}\), Panel A. The cavity is lined with positively charged amino acids, though charge interactions are likely not sufficient to guide NTPs into the interior. Indeed, molecular dynamics (MD) simulations have shown that the residues of the NTP channel can also explore a relatively large amount of space, which may allow the RdRp to actively “pump” NTPs down the channel.
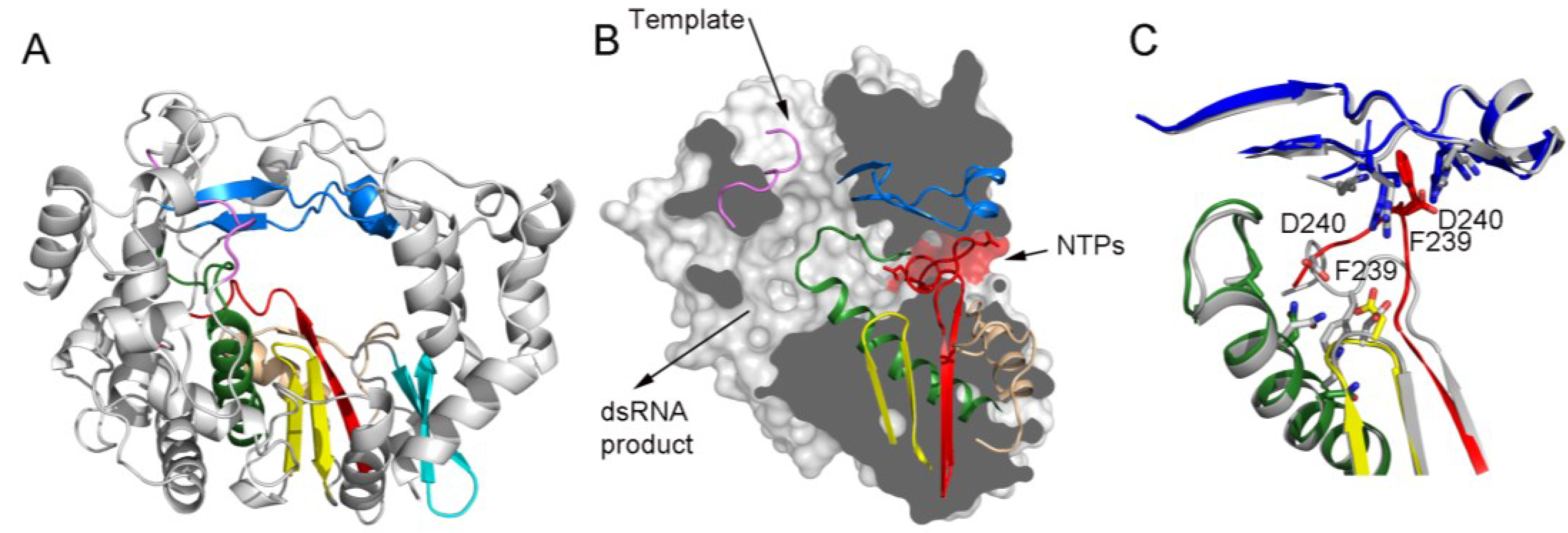
Panel (A) shows a ribbon representation of a typical picornaviral RdRP (model from the cardiovirus EMCV 3Dpol, PDB id. 4NZ0). The seven conserved motifs are indicated in different colors: motif A, red; motif B, green; motif C, yellow; motif D, sand; motif E, cyan; motif F, blue; motif G, pink;
Panel (B) shows a lateral view of a surface representation of the enzyme (grey) that has been cut to expose the three channels that are the entry and exit sites of the different substrates and reaction products. The structural elements that support motifs A–G are also shown as ribbons. This panel also shows the organization of the palm sub-domain with motif A shown in two alternative conformations: the standard conformation (PDB id. 4NZ0) found in the apo-form of most crystallized 3Dpol proteins and the altered conformation found in the tetragonal crystal form of the EMCV enzyme (PDB id. 4NYZ). The alterations affect mainly Asp240, the amino acid in charge of incoming ribonucleotide triphosphate (rNTP) selection, and the neighboring Phe239 that move ~10 Å away from its position in the enzyme catalytic cavity directed towards the entrance of the nucleotide channel, approaching to motif F;
Panel (C) shows a close-up of the structural superimposition of the two alternative conformations of the EMCV motif A;
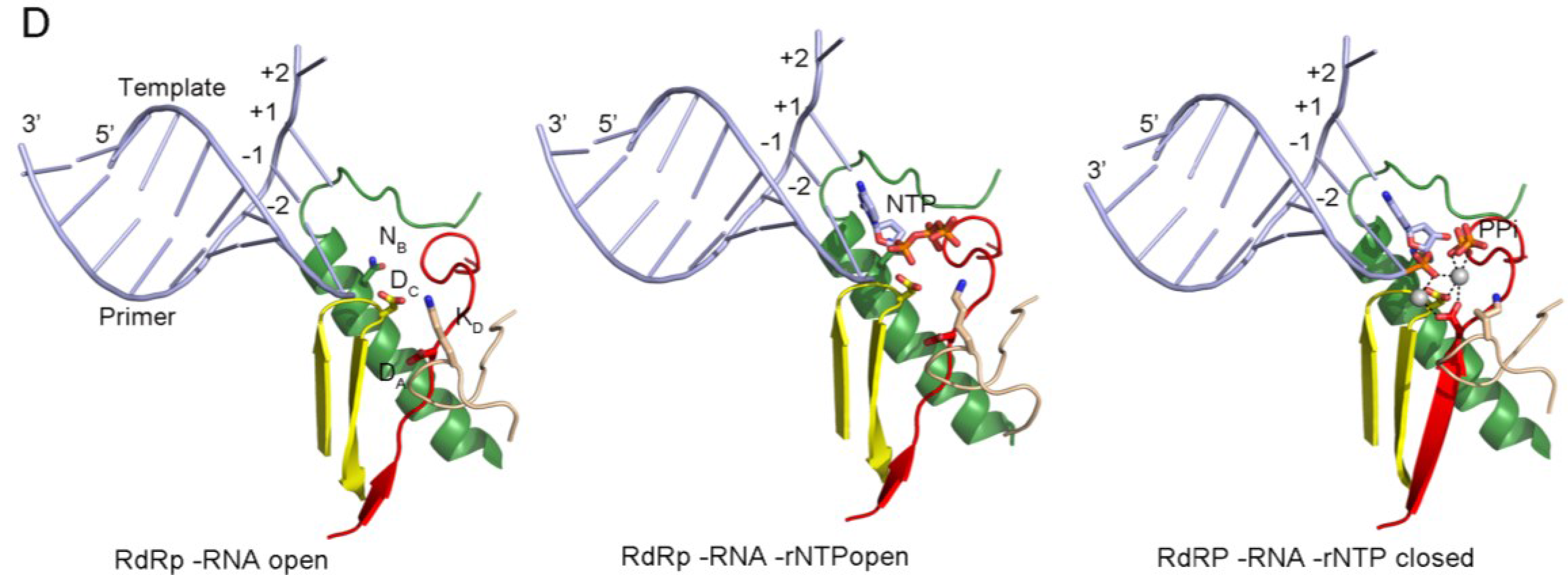
Panel (D) shows the PV replication-elongation complexes. Sequential structures illustrating the movement of the different palm residues from a binary PV 3Dpol-RNA open complex (left) to an open 3Dpol-RNA-rNTP ternary complex (middle) where the incoming rNTP is positioned in the active site for catalysis and, a closed ternary complex (right) after nucleotide incorporation and pyrophosphate (PPi) release. The residues DA (involved in rNTP selection through an interaction with the 2′ hydroxyl group), DC (the catalytic aspartate of motif C), KD (the general acid residue of motif D that can coordinate the export of the PPi group), and NB (a conserved Asn of motif B, interacting with DA) have been highlighted as sticks. The different structures correspond to the 3Dpol-RNA (PDB id. 3OL6), 3Dpol-RNA-CTP open complex (PDB id. 3OLB) and 3Dpol-RNA-CTP closed complex (PDB id. 3OL7) structures of PV elongation complexes, respectively
The finger subdomain residues mainly pack into the major groove of the RNA template. Furthermore, upon entry of the template, they can unstack the single strand at position +3, as shown in Figure \(\PageIndex{5}\) (Panel A). The non-conserved structural elements of the fingers subdomain play a role in RNA binding as well. In particular, the fingertips of dsRNA and some +RNA virus RdRps allow the polymerase to “hold” the template without extensive conformational changes. The variations and extensions in the fingers subdomain has also been shown to play roles in protein–protein interactions, phosphorylation, oligomerization, and nuclear import. In contrast, the HIV-1 RT lacks such extensions and adopts a more “open-hand” or U-shaped binding cleft. As a consequence, the RT structure must rearrange its subdomains to coordinate the binding of the template, nascent strand, and incoming dNTPs.
Panel a shows the structure of the PV active site as it moves from a native state or elongation complex (i) to an open complex (ii), and a closed complex (iii). The closed complex depicted here shows the active site after catalysis. Highlighted are the metal-binding aspartates of motifs A and C, and the lysine of motif D that acts as a general acid. Color coding by motif as in Fig. 26.3.2. Image based on PDB accessions 3Ol6, 3OLB, and 3OL7.
Panel b shows a schematic presentation of the RdRp active site. The aspartates (Asp) of motif A (yellow) and C (orange) bind divalent metal ions (marked Mg and shaded grey), which are used to coordinate the formation of a new phosphodiester bond at the 3′-OH (red in panel ii) of the nascent strand (yellow). The general acid (red Lys/His in panel ii) is positioned near the β-phosphate of the incoming NTP to protonate the PPi leaving group.
Panel c shows a simplified schematic of the kinetic steps of RNA polymerases. Asterisk indicates a closed complex
The NTP and template entry channels meet at the palm subdomain - Figure \(\PageIndex{5}\), Panel A. This is a structure that is comprised of a central, partially formed three-stranded β-sheet, which is also present in RNA-recognition motifs (RRMs). The relative movement of these strands within the RRM is vital to catalysis and dependent on NTP binding. Only when a correct NTP binds can motif A and motif C align and the RRM fold be completed. This catalytically competent conformation of the active site is often referred to as the closed complex (not to be confused with the “closed-hand”, which refers to the overall structure of the RdRp) - Figure \(\PageIndex{5}\), Panel d.
The polymerase reaction creates new phosphodiester bonds between NTPs using RNA as a template. The NTP substrates involved in this reaction are coordinated by two metal ions, which are bound by the conserved aspartates of motifs A and C - Figure \(\PageIndex{5}\) - panel a, i. They also position the NTP’s triphosphate optimally for attack by the sugar moiety of the nascent strand once its 3′ carbon has lost a proton- Figure \(\PageIndex{5}\), panel b. The N-terminal aspartate of motif C thus uses a metal ion to fix the α-phosphate of the incoming NTP and reduce the pKa of the nascent RNA’s 3′-OH to facilitate the attack - Figure \(\PageIndex{5}\), panel b, ii. The amino-terminal carboxylate of motif A, on the other hand, stabilizes the β- and γ-phosphates with its divalent metal as well as the pentavalent (phosphorane) intermediate that forms during catalysis - Figure \(\PageIndex{5}\), panel b, ii. Structural and biochemical analyses have shown that the formation of this transient structure is dependent on the attack of the NTP’s α-phosphate by the 3′-OH, which is often the rate-limiting catalytic step in NTP condensation - Figure \(\PageIndex{5}\), panel b.
Motif D’s lysine or histidine assists the N-terminal aspartate of motif A in coordinating the β-phosphate of the incoming NTP, analogous to the trigger loop in DdDps. However, when the positively charged side chain of motif D approaches the β-phosphate, it can protonate the PPi leaving group as well - Figure \(\PageIndex{5}\), panel b, ii. This second protonation step in the active site is not essential for the polymerase reaction, since catalysis can still take place when motif D’s lysine has been mutated to a residue with a different pKa. The polymerase reaction rate will nevertheless be 50- to 1,000-fold higher when a lysine or histidine is present. Recent data even suggests that motif D can coordinate the export of the PPi group from the active site once catalysis has taken place, thereby triggering the translocation of the RNA.
Overall, the RNA replication process can be summarized with this four-step mechanism:
- Nucleoside triphosphate (NTP) binding - initially, the RdRp presents with a vacant active site in which an NTP binds, complementary to the corresponding nucleotide on the template strand. Correct NTP binding causes the RdRp to undergo a conformational change.
- Active site closure - the conformational change, initiated by the correct NTP binding, results in the restriction of active site access and produces a catalytically competent state.
- Phosphodiester bond formation - two Mg2+ ions are present in the catalytically active state and arrange themselves in such a way around the newly synthesized RNA chain that the substrate NTP can undergo a phosphatidyl transfer and form a phosphodiester bond with the newly synthesized chain. With the use of these Mg2+ ions, the active site is no longer catalytically stable, and the RdRp complex changes to an open conformation.
- Translocation - once the active site is open, the RNA template strand can move by one position through the RdRp protein complex and continue chain elongation by binding a new NTP, unless otherwise specified by the template.
Figure \(\PageIndex{6}\) shows a more detailed representation of the elongation catalytic cycle.
Figure \(\PageIndex{6}\): Elongation catalytic cycle of RNA-dependent RNA polymerases
RNA synthesis can be performed using a primer-independent (de novo) or a primer-dependent mechanism that utilizes a viral protein genome-linked (VPg) primer. The de novo initiation consists of the addition of a nucleoside triphosphate (NTP) to the 3'-OH of the first initiating NTP. During the following so-called elongation phase, this nucleotidyl transfer reaction is repeated with subsequent NTPs to generate the complementary RNA product. The termination of the nascent RNA chain produced by RdRp is not completely known, however, it has been shown that RdRp termination is sequence-independent.
One feature of RNA-dependent RNA polymerase replication is the immense error rate during transcription. RdRps and RTs are known to have a lack of fidelity on the order of 104 nucleotides, which is thought to be a direct result of their insufficient proofreading abilities. This high rate of variation is favored in viral genomes as it allows for the pathogen to overcome defenses developed by hosts trying to avoid infection allowing for evolutionary growth.
Let's look at another RdRp from the poliovirus. The virus has a single-stranded, positive-sense RNA genome, which makes it infectious in itself but much more infectious when replicated. It is translated into a polyprotein which is cleaved into about 12 separate proteins. One protein, 3Dpol, is an RNA-dependent RNA polymerase that transcribes the infecting +RNA strand into a -RNA strand, which then serves as a template for more +RNA strands.
Figure \(\PageIndex{7}\) shows an interactive iCn3D model of the Poliovirus polymerase elongation complex with 2',3'-dideoxy-CTP (3OLB). The model shows the palm, finger, and thumb domains.
Color coding is as follows:
- red: finger domain
- pink: pinky finger part of the finger domain
- blue: thumb domain
- light blue: primer grip at the beginning of the thumb domain
- cyan: RNA template
- green: RNA product
- gray: palm domain
Structural studies suggest that the pinky finger is involved in the initiation, while nucleotide binding and catalysis used the palm domain. The thumb domain appears to affect the translocation step. In most nucleases, the finger binds and positions NTPs in the active site. After catalysis, the reverse motion opens the active site which allows translocation by effectively ratcheting the template by one base pair which is driven by PPi release. These conformational changes can't be made so easily in +RNA stranded viral RdRps.
References
1. te Velthuis, A.J.W. (2014) Common and unique features of viral RNA-dependent polymerases. Cell. & Mol Life Sci 71:4403-4420. Available at: https://link.springer.com/article/10.1007/s00018-014-1695-z#Sec1
2. Wikipedia contributors. (2021, August 2). RNA-dependent RNA polymerase. In Wikipedia, The Free Encyclopedia. Retrieved 02:47, August 8, 2021, from https://en.Wikipedia.org/w/index.php?title=RNA-dependent_RNA_polymerase&oldid=1036813791
3. Ferrer-Orta, C., Ferrero, D., and Verdaguer, N. (2015) RNA-Dependent RNA Polymerases of Picornaviruses: From the Structure to Regulatory Mechanisms (2015 Viruses 7(8):4438-4460. Available at: https://www.mdpi.com/1999-4915/7/8/2829/htm




_.png?revision=1&size=bestfit&width=521&height=414)
.png?revision=1&size=bestfit&width=425&height=413)