17.2: Membrane Transport
- Page ID
- 89008
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The first control on the passage of molecules across membranes is the semipermeable character of the membrane itself. Molecules move in and out of cells in one of three ways: passive diffusion, facilitated diffusion, and active transport.
Only a few small, relatively uncharged molecules can cross a membrane unassisted (i.e., by passive diffusion). Hydrophilic molecules that must enter or leave cells require help (i.e., from facilitated diffusion). Passive and facilitated diffusion release the free energy inherent in concentration gradients as molecules diffuse across a membrane.
In contrast, active transport (i.e., membrane pumps) consumes energy to create concentration gradients of specific solutes. The specificity of facilitated diffusion and active transport lies in the integral membrane proteins that recognize and bind specific solutes for transport. As you may predict, allosteric regulation of these proteins controls the movement of their target molecules into or out of cells.
Despite water’s polarity, many believed that the small water molecules crossed membranes without help. Indeed, they do to a limited extent. However, others suspected that given its highly charged polar covalent bonds relative to its small size, a water molecule would require an assist to get across membranes efficiently. Let’s begin with a closer look at passive diffusion and diffusion by facilitated diffusion, followed by osmosis (a special case of facilitated diffusion of water), and finally active transport.
17.2.1 Passive Diffusion of Solutes
Diffusion across membranes does not require energy. In fact, diffusion to relieve a solute concentration gradient can release energy—recall the movement of protons through the F1 ATP synthase proton gate that synthesizes ATP during mitochondrial oxidative phosphorylation. Passive diffusion in solution is the movement of molecules over time by random motion (also called Brownian motion) from regions of higher to regions of lower concentration. Significant passive diffusion across cellular membranes is limited to a few molecules, mostly gases like \(\rm O_2\), \(\rm CO_2\), and \(\rm N_2\), that can freely cross the hydrophobic phospholipid barrier. The rapid diffusion of gases is essential for \(\rm O_2\) and \(CO_2\) exchange between the alveolar capillaries and the cells of the lungs during physiological respiration. \(\rm O_2\) and \(\rm CO_2\) exchange also occurs in mitochondria during cellular respiration. The rate of diffusion of a molecule is dependent only on its own concentration. It is unaffected by the concentration of other molecules. Over time, the random motion of solutes within and across compartments results in a dynamic equilibrium for each different solute. At equilibrium, solute molecules continue to diffuse across the membrane, but for each molecule moving across in one direction, another molecule of the same solute crosses in the other direction.
17.2.2 Facilitated Diffusion of Solutes and Ions
Like the passive diffusion of those gasses, facilitated diffusion (e.g., of ATP through an F1 ATP synthase) is the spontaneous (downhill) passage of molecules or ions across membranes, but with the help of specific transmembrane proteins. The kinetics of passive and facilitated diffusion reveals the differences between the two processes. To understand the latter, recall that the rate of enzyme catalysis is saturable. That is, as the concentration of substrate is increased, the rate of the catalyzed reaction approaches a maximum (Vmax). This occurs when all enzyme molecules in solution are bound to substrate molecules. The same saturation phenomenon applies to facilitated diffusion—the rate of diffusion of a solute across a membrane is directly proportional to the concentration of the solute but is limited by the number of transport proteins in the membrane. Figure 17.1 plots the kinetics of passive and facilitated diffusion.

Perhaps you see another similarity between facilitated diffusion and enzyme catalysis in this graph! Relative rates of facilitated diffusion are typically rapid, compared to those of passive diffusion. This is because the allosteric changes that accompany facilitated diffusion are rapid, just as they are during enzyme catalysis. This should suggest yet another similarity between enzymatic catalysis and facilitated transport: both can be regulated!
Figure 17.2 illustrates three kinds of facilitated transport of solutes.
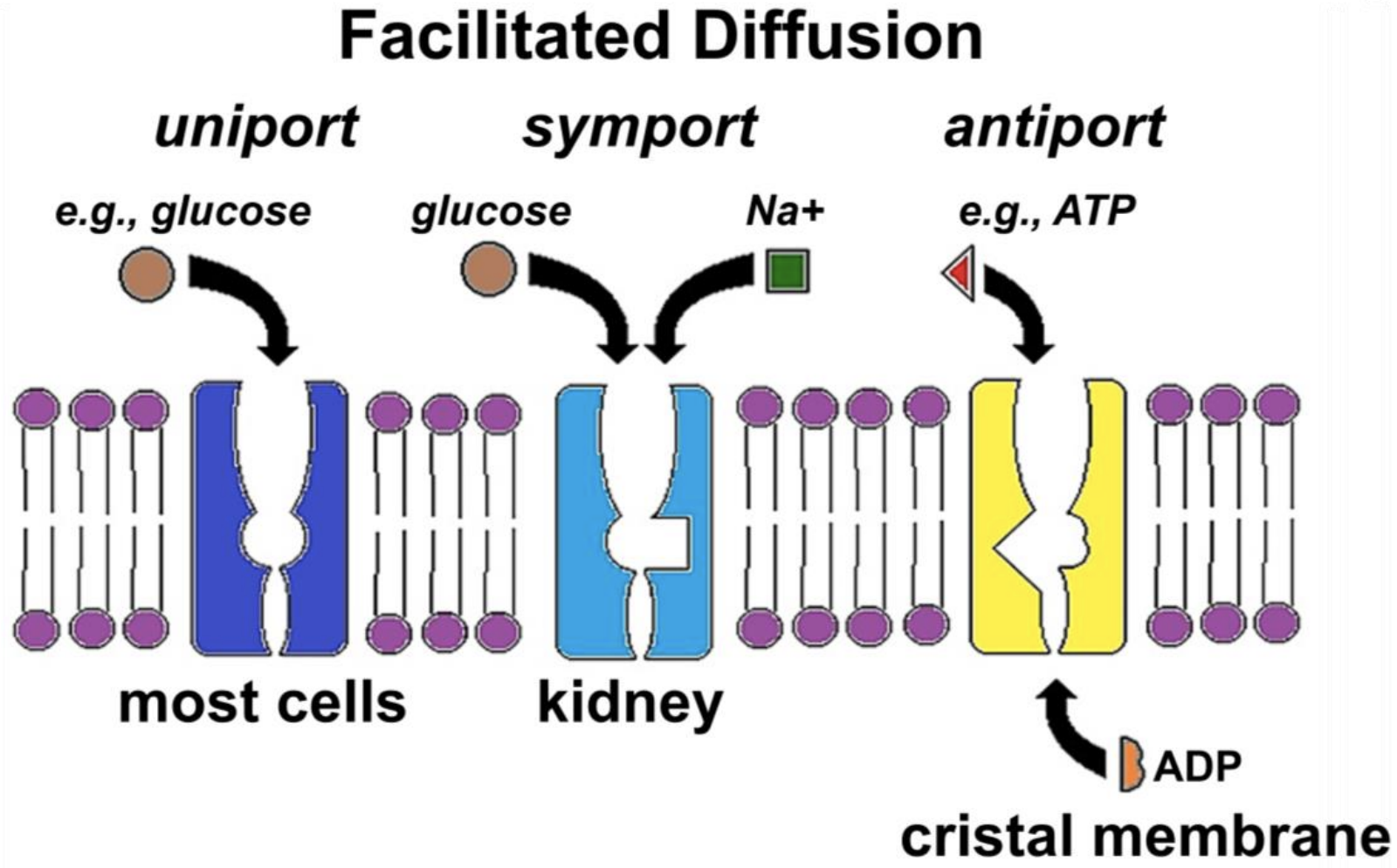
The GLUT (GLUcose Transporter) protein (Figure 17.2, left) allows glucose uniport, the specific transport of a single substance in or out of cells. In symport (Figure 17.2, middle) glucose transporters couple the simultaneous movement of glucose and sodium ions (e.g., in kidney cells). The SGLT (Sodium-GLucose Transporter) serves a similar function in small intestine cells, enabling absorption of dietary glucose and sodium. Antiport (Figure 17.2, right) allows the specific exchange of molecules across a membrane. In the example shown, ATP leaves the mitochondrial matrix, crossing the cristal membrane at the same time as ADP enters the matrix. Whether by uniport, symport, or antiport, each solute will independently cross a membrane down its concentration gradient, moving from regions where it is at a higher concentration to regions of lower concentration. Recall that the direction of diffusion depends on relative concentrations of the solutes, and that diffusion along (i.e., down) a gradient releases free energy.
Each of the examples in Figure 17.2 is accompanied by allosteric changes in a transporter. Model the allosteric changes needed to complete an act of symport or antiport and state your hypothesis!
Proteins mediating facilitated diffusion are of two kinds: carrier proteins and channel proteins. Carrier proteins allow diffusion of large solutes across the membrane. While small, ions also need help to cross the hydrophobic membrane barrier because they have a high charge-to-mass ratio. This is the job of channel proteins, which essentially serve as ion pores.
Like all transporter proteins, both carrier and channel proteins undergo allosteric change during transport. They are also typically subject to allosteric regulation, rather than being in a constant “open” state. Examples of facilitated diffusion are considered in more detail next.
17.2.2.a Carrier Proteins
When a carrier protein binds a solute that must cross the membrane, it undergoes the first of a series of allosteric changes, as shown in Figure 17.3.

During transport itself, the carrier protein undergoes another change in shape. When the solute reaches the other side of the membrane, it no longer has a high affinity for the carrier protein. After release of the solute, a final allosteric change restores the original conformation of the transport protein.
A given carrier protein is specific for a single solute, or at most a single family of closely related solutes. Thus, GLUT1, a glucose uniport transporter, allows glucose (but not fructose or ribose!) to cross membranes. Other specific carrier proteins facilitate the transport of amino acids or charged solutes across cell membranes. Once again, molecules that indicate cell status (i.e., a need to import or export solute) are allosteric effectors that regulate carrier proteins. Insulin is a perfect example of the regulation of solute transport, specifically glucose transport into cells. One consequence of insulin being released during a meal (or in anticipation of a meal!) is the stimulation of glucose transporters to take up glucose. An inability of those transporters to respond to insulin accounts, in part for type-2 (adult-onset) diabetes.
Water gets across membranes by osmosis; we’ll look more closely at how osmosis affects cells in a moment. But first, recall that small amounts of water can cross the phospholipid bilayer unassisted. Water can also cross a membrane incidentally when ions flow through their channel proteins. But most osmosis involves facilitated diffusion mediated by aquaporins. Some aquaporins only transport water. Others have evolved to co-facilitate the transport of glucose (see above), glycerol, urea, ammonia, carbon dioxide, and even ions (protons) along with water.
Like other carrier proteins, aquaporins are allosterically regulated to allow cells to meet specific water balance requirements. So fundamental was the understanding of water balance that the discovery of aquaporins earned Peter Agre a Nobel Prize in Chemistry in 2003.
Since Agre’s discovery (in 1992), several genetic diseases have been linked to aquaporin gene mutations. Kidney cells are critically involved in vertebrate water balance and have many aquaporins in their membranes. In a rare form of diabetes, abnormal aquaporins cause the kidneys to excrete unusually large volumes of water. In another example, aquaporin gene mutations lead to the development of cataracts in both eyes. Since their initial discovery, aquaporins have also been described in bacteria and plants. To learn more, see Aquaporins.
17.2.2.b Ion Channels
Allosteric regulation of ion channel proteins controls ion homeostasis in blood and extracellular fluids within narrow limits. Often, multiple integral proteins contribute to the formation of an ion channel. When stimulated, channel proteins rearrange to open a pore, allowing specific ion transport. Some ion channels, like the glucose-sodium ion symport system noted earlier, mobilize the energy of diffusion of one solute (the ion in this case) to rapidly transport another solute through the same channel (acting like an ion channel and a carrier protein). Finally, ion channels are responsible for the excitability of cells, where \(\rm Na^{+}\), \(\rm K^{+}\), and \(\rm Ca^{++}\) channels collaborate in ion movements into and out of cells leading to neuronal or muscle cell responses (more shortly!).


