B2. Lysozyme
- Page ID
- 5538
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
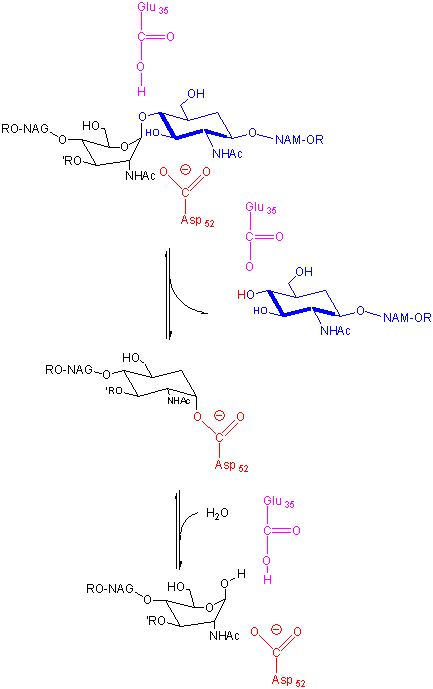
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The lysozyme enzyme, found in cells and secretions of vertebrates but also in viruses which infect bacteria, cleaves peptidoglycan GlcNAc (β 1->4) MurNAc repeat linkages (NAG-NAM) in the cell walls of bacteria and the GlcNAc (β 1->4) GlcNAc (poly-NAG) in chitin, found in the cells walls of certain fungi. Since these polymers are hydrophilic, the active site of the enzyme would be expected to contain a solvent-accessible channel into which the polymer could bind. The crystal structures of lysozyme and complexes of lysozyme and NAG have been solved to high resolution. The inhibitors and substrates form strong H bonds and some hydrophobic interactions with the enzyme cleft. Kinetic studies using (NAG)n polymers show a sharp increase in kcat as n increases from 4 to 5. The kcat for NAG6 and (NAG-NAM)3 are similar. Models studies have shown that for catalysis to occur, (NAG-NAM)3 binds to the active site with each sugar in the chair conformation except the fourth which is distorted to a half chair form, which labilizes the glycosidic link between the 4th and 5th sugars. Additional studies show that if the sugars that fit into the binding site are labeled A-F, then because of the bulky lactyl substituent on the NAM, residues C and E can not be NAM, which suggests that B, D and F must be NAM residues. Cleavage occurs between residues D and E.
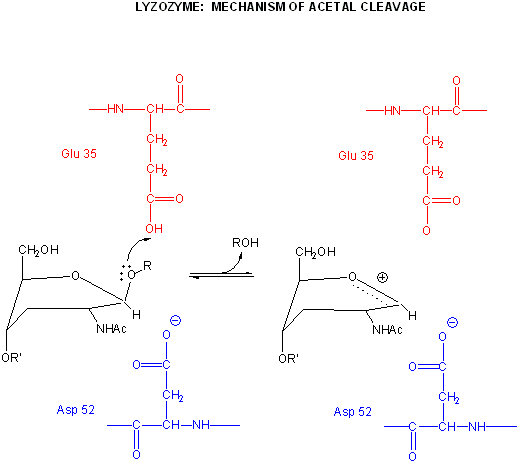
A review of the chemistry of glycosidic bond (an acetal) formation and cleavage shows the acetal cleavage is catalyzed by acids and proceeds by way of an oxonium ion which exists in resonance form as a carbocation.

Catalysis by the enzyme involves Glu 35 and Asp 52 which are in the active site. Asp 52 is surrounded by polar groups but Glu 35 is in a hydrophobic environment. This should increase the apparent pKa of Glu 35, making it less likely to donate a proton and acquire a negative charge at low pH values, making it a better general acid at higher pH values. The general mechanism appears to involve:
- binding of a hexasaccharide unit of the peptidoglycan with concomitant distortion of the D NAM.
- protonation of the sessile acetal O by the general acid Glu 35 (with the elevated pKa), which facilitates cleavage of the glycosidic link and formation of the resonant stabilized oxonium ion.
- Asp 52 stabilizes the positive oxonium through electrostatic catalysis. The distorted half-chair form of the D NAM stabilizes the oxonium which requires co-planarity of the substituents attached to the sp2 hybridized carbon of the carbocation resonant form (much like we saw with the planar peptide bond).
- water attacks the stablized carbocation, forming the hemiacetal with release of the extra proton from water to the deprotonated Glu 35 reforming the general acid catalysis.
Binding and distortion of the D substituent of the substrate (to the half chair form as shown above) occurs before catalysis. Since this distortion helps stabilize the oxonium ion intermediate, it presumably stabilizes the transition state as well. Hence this enzyme appears to bind the transition state more tightly than the free, undistorted substrate, which is yet another method of catalysis.
pH studies show that side chains with pKa's of 3.5 and 6.3 are required for activity. These presumably correspond to Asp 52 and Glu 35, respectively. If the carboxy groups of lysozyme are chemically modified in the presence of a competitive inhibitor of the enzyme, the only protected carboxy groups are Asp 52 and Glu 35.
In an alternative mechanism, Asp 52 acts as a nucleophilic catalysis and forms a covalent bond with NAM, expelling a NAG leaving group with Glu 35 acting as a general acid. This alternative mechanism also is consistent with other β-glycosidic bond cleavage enzyme. Substrate distortion is also important in this alternative mechanism.
Figure: alternative mechanism