Gene regulation: Bacterial*#
( \newcommand{\kernel}{\mathrm{null}\,}\)
Examples of Bacterial Gene Regulation
This section describes two examples of transcriptional regulation in bacteria. These are presented as illustrative examples. Use these examples to learn some basic principles about mechanisms of transcriptional regulation. Be on the lookout in class, in discussion, and in the study-guides for extensions of these ideas and use these to explain the regulatory mechanisms used for regulating other genes.
Gene Regulation Examples in E. coli
The DNA of bacteria and archaea are usually organized into one or more circular chromosomes in the cytoplasm. The dense aggregate of DNA that can be seen in electron micrographs is called the nucleoid. In bacteria and archaea, genes, whose expression needs to be tightly coordinated (e.g. genes encoding proteins that are involved in the same biochemical pathway) are often grouped closely together in the genome. When the expression of multiple genes is controlled by the same promoter and a single transcript is produced these expression units are called operons. For example, in the bacterium Escherschia coli all of the genes needed to utilize lactose are encoded next to one another in the genome. This arrangement is called the lactose (or lac) operon. It is often the case in bacteria and archaea that nearly 50% of all genes are encoded into operons of two or more genes.
The Role of the Promoter
The first level of control of gene expression is at the promoter itself. Some promoters recruit RNA polymerase and turn those DNA-protein binding events into transcripts more efficiently than other promoters. This intrinsic property of a promoter, it's ability to produce transcript at a particular rate, is referred to as promoter strength. The stronger the promoter, the more RNA is made in any given time period. Promoter strength can be "tuned" by Nature in very small or very large steps by changing the nucleotide sequence the promoter (e.g. mutating the promoter). This results in families of promoters with different strengths that can be used to control the maximum rate of gene expression for certain genes.
UC Davis Undergraduate Connection:
A group of UC Davis students interested in synthetic biology used this idea to create synthetic promoter libraries for engineering microbes as part of their design project for the 2011 iGEM competition.
Example #1: Trp Operon
Logic for regulating tryptophan biosynthesis
E. coli, like all organisms, needs to either synthesize or consume amino acids to survive. The amino acid tryptophan is one such amino acid. E. colican either import tryptophan from the environment (eating what it can scavenge from the world around it) or synthesize tryptophan de novo using enzymes that are encoded by five genes. These five genes are encoded next to each other in the E. coli genome into what is called the tryptophan (trp) operon (Figure below). If tryptophan is present in the environment, then E. coli does not need to synthesize it and the switch controlling the activation of the genes in the trp operon is switched off. However, when environmental tryptophan availability is low, the switch controlling the operon is turned on, transcription is initiated, the genes are expressed, and tryptophan is synthesized. See the figure and paragraphs below for a mechanistic explanation.
Organization of the trp operon
Five genomic regions encoding tryptophan biosynthesis enzymes are arranged sequentially on the chromosome and are under the control of a single promoter - they are organized into an operon. Just before the coding region is the transcriptional start site. This is, as the name implies, the location where the RNA polymerase starts a new transcript. The promoter sequence is further upstream of the transcriptional start site.
A DNA sequence called an "operator" is also encoded between the promoter and the first trp coding gene. This operator is the DNA sequence to which the transcription factor protein will bind.
A few more details regarding TF binding sites
It should be noted that the use of the term "operator" is limited to just a few regulatory systems and almost always refers to the binding site for a negatively acting transcription factor. Conceptually what you need to remember is that there are sites on the DNA that interact with regulatory proteins allowing them to perform their appropriate function (e.g. repress or activate transcription). This theme will be repeated universally across biology whether the "operator" term is used or not.
Moreover, while the specific examples you will be show depict TF binding sites in their known locations, these locations are not universal to all systems. Transcription factor binding sites can vary in location relative to the promoter. There are some patterns (e.g. positive regulators are often upstream of the promoter and negative regulators bind downstream), but these generalizations are not true for all cases. Again, the key thing to remember is that transcription factors (both positive and negatively acting) have binding sites with which they interact to help regulate the initiation of transcription by RNA polymerase.
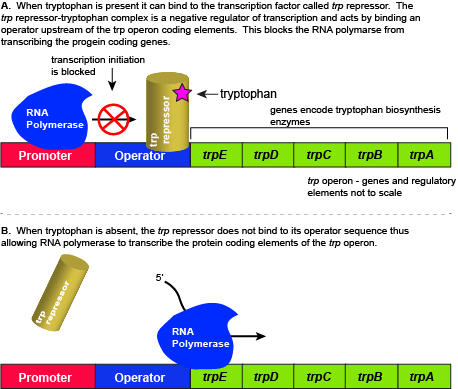
The five genes that are needed to synthesize tryptophan in E. coli are located next to each other in the trp operon. When tryptophan is plentiful, two tryptophan molecules bind to the transcription factor and allow the TF-tryptophan complex to bind at the operator sequence. This physically blocks the RNA polymerase from transcribing the tryptophan biosynthesis genes. When tryptophan is absent, the transcription factor does not bind to the operator and the genes are transcribed.
Attribution: Marc T. Facciotti (own work)
Regulation of the trp operon
When tryptophan is present in the cell: two tryptophan molecules bind to the trp repressor protein. When tryptophan binds to the transcription factor it causes a conformational change in the protein which now allows the TF-tryptophan complex to bind to the trp operator sequence. Binding of the tryptophan–repressor complex at the operator physically prevents the RNA polymerase from binding, and transcribing the downstream genes. When tryptophan is not present in the cell, the transcription factor does not bind to the operator; therefore, the transcription proceeds, the tryptophan utilization genes are transcribed and translated, and tryptophan is thus synthesized.
Since the transcription factor actively binds to the operator to keep the genes turned off, the trp operon is said to be "negatively regulated" and the proteins that bind to the operator to silence trp expression are negative regulators.
Suggested discussion
Do you think that the constitutive expression levels of the trp operon are high or low? Why?
Suggestion discussion
Suppose nature took a different approach to regulating the trp operon. Design a method for regulating the expression of the trp operon with a positive regulator instead of a negative regulator. (hint: we ask this kind of question all of the time on exams)
External link
Watch this video to learn more about the trp operon.
Example #2: The lac operon
Rationale for studying the lac operon
In this example, we examine the regulation of genes encoding proteins whose physiological role is to import and assimilate the disaccharide lactose, the lac operon. The story of the regulation of lac operon is a common example used in many introductory biology classes to illustrate basic principles of inducible gene regulation. We choose to describe this example second because it is, in our estimation, more complicated than the previous example involving the activity of a single negatively acting transcription factor. By contrast, the regulation of the lac operon is, in our opinion, a wonderful example of how the coordinated activity of both positive and negative regulators around the same promoter can be used to integrate multiple different sources of cellular information to regulate the expression of genes.
As you go through this example, keep in mind the last point. For many Bis2a instructors it is more important for you to learn the lac operon story than it is to know the logic table presented below. For those instructors for which this is the case, they will usually make a point to let you know and often deliberately not include exam questions about the lac operon. Rather they will test you on whether you understood the basis underlying the regulatory mechanisms that you study. If it's not clear what the instructor wants you should ask.
The utilization of lactose
Lactose is a disaccharide composed of the hexoses glucose and galactose. It is commonly found in high abundance in milk and some milk products. As one can imagine, the disaccharide can be an important food-stuff for microbes that are able to utilize its two hexoses. E. coli is able to use multiple different sugars as energy and carbon sources, including lactose and the lac operon is a structure that encodes the genes necessary to acquire and process lactose from the local environment. Lactose, however, has not been frequently encountered by E. coli during its evolution and therefore the genes of the lac operon must typically be repressed (i.e. "turned off") when lactose is absent. Driving transcription of these genes when lactose is absent would waste precious cellular energy. By contrast, when lactose is present, it would make logical sense for the genes responsible for the utilization of the sugar to be expressed (i.e. "turned on"). So far the story is very similar to that of the tryptophan operon described above.
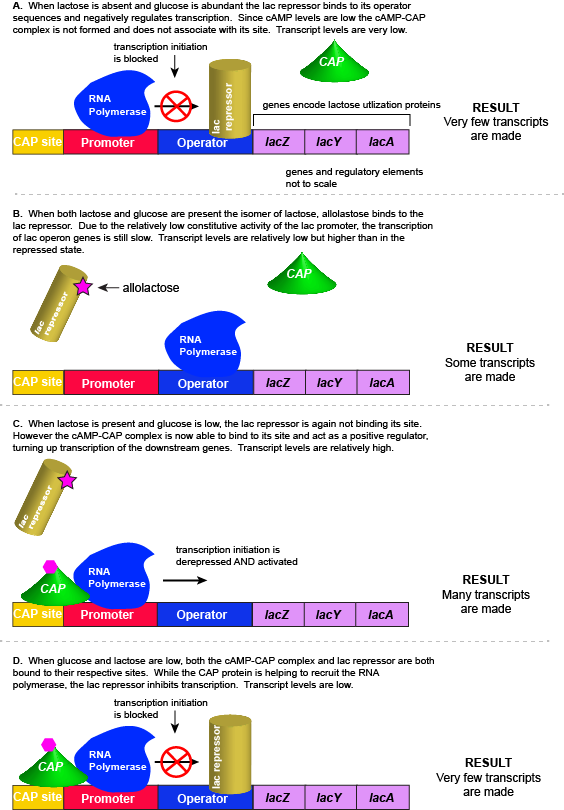
However, there is a catch. Experiments conducted in the 1950's by Jacob and Monod clearly demonstrated that E. coli prefers to utilize all the glucose present in the environment before it begins to utilize lactose. This means that the mechanism used to decide whether or not to express the lactose utilization genes must be able to integrate two types of information (1) the concentration of glucose and (2) the concentration of lactose. While this could theoretically be accomplished in multiple ways, we will examine how the lac operon accomplishes this by using multiple transcription factors.
The transcriptional regulators of the lac operon
The lac repressor - a direct sensor of lactose
As noted, the lac operon normally has very low to no transcriptional output in the absence of lactose. This is due to two factors: (1) the constitutive promoter strength for the operon is relatively low and (2) the constant presence of the LacI repressor protein negatively influences transcription. This protein binds to the operator site near the promoter and blocks RNA polymerase from transcribing the lac operon genes. By contrast, if lactose is present, lactose will bind to the LacI protein, inducing a conformational change that prevents LacI-lactose complex from binding to its binding sites. Therefore, when lactose is present the negative regulatory LacI is not bound to the its binding site and transcription of lactose utilizing genes can proceed.
CAP protein - an indirect sensor of glucose
In E. coli, when glucose levels drop, the small molecule cyclic AMP (cAMP) begins to accumulate in the cell. cAMP is a common signaling molecule that is involved in glucose and energy metabolism in many organisms. When glucose levels decline in the cell, the increasing concentrations of cAMP allow this compound to bind to the positive transcriptional regulator called catabolite activator protein (CAP) - also referred to as CRP. cAMP-CAP complex has many sites located throughout the E. coli genome and many of these sites are located near the promoters of many operons that control the processing of various sugars.
In the lac operon, the cAMP-CAP binding site is located upstream of the promoter. Binding of cAMP-CAP to the DNA helps to recruit and retain RNA polymerase to the promoter. The increased occupancy of RNA polymerase to its promoter, in turn, results in increased transcriptional output. In this case the CAP protein is acting as a positive regulator.
Note that the CAP-cAMP complex can, in other operons, also act as a negative regulator depending upon where the binding site for CAP-cAMP complex is located relative to the RNA polymerase binding site.
Putting it all together: Inducing expression of the lac operon
For the lac operon to be activated, two conditions must be met. First, the level of glucose must be very low or non-existent. Second, lactose must be present. Only when glucose is absent and lactose is present will the lac operon be transcribed. When this condition is achieved the LacI-lactose complex dissociates the negative regulator from near the promoter, freeing the RNA polymerase to transcribe the operon's genes. Moreover, high cAMP (indirectly indicative of low glucose) levels trigger the formation of the CAP-cAMP complex. This TF-inducer pair now bind near the promoter and act to positively recruit the RNA polymerase. This added positive influence boosts transcriptional output and lactose can be efficiently utilized. The mechanistic output of other combinations of binary glucose and lactose conditions are descried in the table below and in the figure that follows.
A more nuanced view of lac repressor function
The description of the lac repressor's function correctly describes the logic of the control mechanism used around the lac promoter. However, the molecular description of binding sites is a bit overly simplified. In reality the lac repressor has three similar, but not identical, binding sites called Operator 1, Operator 2, and Operator 3. Operator 1 is very close to the transcript start site (denoted +1). Operator 2 is located about +400nt into the coding region of the LacZ protein. Operator 3 is located about -80nt before the transcript start site (just "outside" of the CAP binding site).
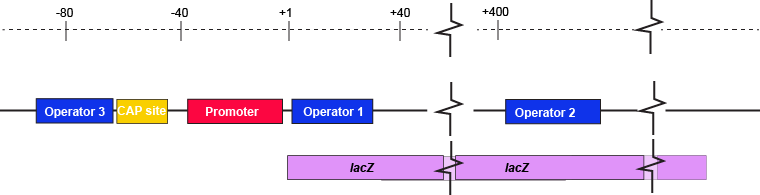
The lac operon regulatory region depicting the promoter, three lac operators, and CAP binding site. The coding region for the Lac Z protein is also shown relative to the operator sequences. Note that two of the operators are in the protein coding region - there are multiple different types of information simultaneously encoded in the DNA.
Attribution: Marc T. Facciotti (own work)
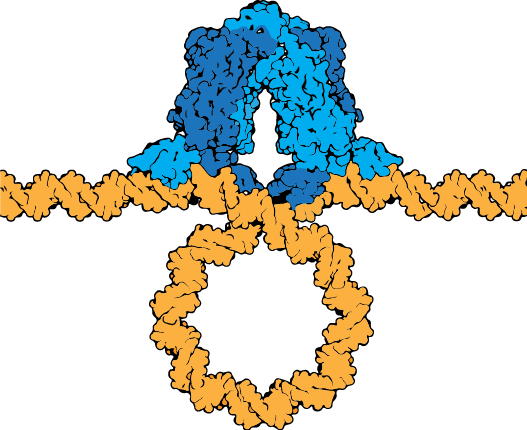
The lac repressor tetramer (blue) depicted binding two operators on a strand of looped DNA (orange).
Attribution: Marc T. Facciotti (own work) - Adapted from Goodsell (https://pdb101.rcsb.org/motm/39)