S2024_Bis2A_Namekawa_DNA_Structure_and_Replication II
- Page ID
- 132377
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
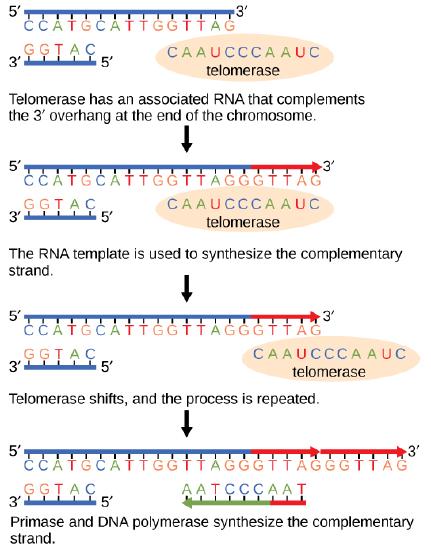
Telomere and telomerase
The ends of replication in circular bacterial chromosomes pose few practical problems. However, the ends of linear eukaryotic chromosomes pose a specific problem for DNA replication. Because DNA polymerase can add nucleotides in only one direction (5' to 3'), the leading strand allows for continuous synthesis until the end of the chromosome is reached; however, as the replication complex arrives at the end of the lagging strand there is no place for the primase to "land" and synthesize an RNA primer so that the synthesis of the missing lagging strand DNA fragment at the end of the chromosome can be initiated by the DNA polymerase. Without some mechanism to help fill this gap, this chromosomal end will remain unpaired and will be lost to nucleases. Over time, and several rounds of replication, this would result in the ends of linear chromosomes getting progressively shorter, ultimately compromising the ability of the organism to survive. These ends of the linear chromosomes are known as telomeres, and nearly all eukaryotic species have evolved repetitive sequences that do not code for a specific gene. As a result, these "non-coding" telomeres act as replication buffers and are shortened with each round of DNA replication instead of critical genes. For example, in humans, a six base-pair sequence, TTAGGG, repeats 100 to 1000 times at the end of most chromosomes. Besides acting as a potential buffer, the discovery of the enzyme telomerase helped in understanding how chromosome ends are maintained. Telomerase is an enzyme composed of protein and RNA. Telomerase attaches to the end of the chromosome by complementary base pairing between the RNA component of telomerase and the DNA template. Telomerase uses the RNA as a complementary strand for the short elongation of its complement. The cell repeats this process many times. Once the lagging strand template is sufficiently elongated by telomerase, primase will create a primer followed by DNA polymerase which can now add nucleotides complementary to the ends of the chromosomes. Thus, the ends of the chromosomes are replicated.
Figure 1. The ends of linear chromosomes are maintained by the action of the telomerase enzyme.
Telomerase is not active in adult somatic cells. Adult somatic cells that undergo cell division continue to have their telomeres shortened. This means that telomere shortening is associated with aging. In 2010, scientists found that telomerase can reverse some age-related conditions in mice, and this may have potential in regenerative medicine. Telomerase-deficient mice were used in these studies; these mice have tissue atrophy, stem-cell depletion, organ system failure, and impaired tissue injury responses. Telomerase reactivation in these mice caused the extension of telomeres, reduced DNA damage, reversed neurodegeneration, and improved functioning of the testes, spleen, and intestines. Thus, telomere reactivation may have potential for treating age-related diseases in humans.
Possible NB Discussion  Point
Point
Imagine that researchers have invented a drug for humans that upregulates telomerase activity. Would you be excited or would you be skeptical about being part of a clinical trial? Do you trust this drug? Can you think of any negative consequences to activating telomerase to a level that it would not “naturally” be at? Assuming scientists proved without doubt that their drug has no negative side effects -- what are some of the ethical questions you would consider?
Replication design challenge: proofreading
When the cell replicates its DNA, it does so in response to environmental signals that tell the cell it is time to divide. The ideal goal of DNA replication is to produce two identical copies of the double-stranded DNA template and to do it in an amount of time that does not pose an unduly high evolutionarily selective cost. This is a daunting task when you consider that there are ~6,500,000,000 base pairs in the human genome and ~4,500,000 base pairs in the genome of a typical E. coli strain and that Nature has determined that the cells must replicate within 24 hours and 20 minutes, respectively. In either case, many individual biochemical reactions need to take place.
While ideally replication would happen with perfect fidelity, DNA replication, like all other biochemical processes, is imperfect—bases may be left out, extra bases may be added, or bases may be added that do not properly base-pair. In many organisms, many of the mistakes that occur during DNA replication are corrected promptly by DNA polymerase itself via a mechanism known as proofreading. In proofreading, the DNA polymerase "reads" each newly added base via sensing the presence or absence of small structural anomalies before adding the next base to the growing strand. In doing so, a correction can be made.
If the polymerase detects that a newly added base has been paired correctly with the base in the template strand, it adds the next nucleotide. If, however, a wrong nucleotide is added to the growing polymer, the misshaped double helix will cause the DNA polymerase to stall, and it will eject the newly made strand from the polymerizing site. Consequently, the DNA strand will enter an exonuclease site. In this site, DNA polymerase can cleave off the last several nucleotides that were added to the polymer. Once the polymerase removes the incorrect nucleotides, the DNA strand can return to the polymerizing site and new nucleotides will be added again. This proofreading capability comes with some trade-offs: using an error-correcting/more accurate polymerase requires time (the trade-off is speed of replication) and energy (always an important cost to consider). The slower you go, the more accurate you can be. Going too slow, however, may keep you from replicating as fast as your competition, so figuring out the balance is key.
We call errors that are not corrected by proofreading mutations.
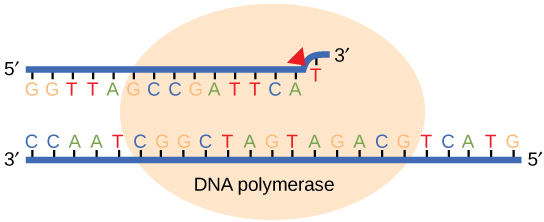
Figure 2. Proofreading by DNA polymerase corrects errors during replication.
Replication mistakes and mismatch repair
Although DNA replication is typically a highly accurate process and proofreading DNA polymerases helps to keep the error rate low, mistakes still occur. Errors not corrected during replication may instead get corrected after replication completes; we know this kind of repair as mismatch repairs. Specific enzymes recognize the incorrectly added nucleotide and excise it, replacing it with the correct base. But, how do mismatch repair enzymes recognize which of the two bases is the incorrect one?
In E. coli, after replication, the nitrogenous base adenine gains a methyl group; this means that directly after replication, the parental DNA strand will have methyl groups, whereas the newly synthesized strand lacks them. Thus, mismatch repair enzymes can scan the DNA and remove the wrongly incorporated bases from the newly synthesized, non-methylated strand by using the methylated strand as the "correct" template from which to incorporate a new nucleotide. In eukaryotes, the mechanism is not as well understood, but we believe it to involve recognition of unsealed nicks in the new strand, and a short-term, continuing association of some replication proteins with the new daughter strand after replication has completed. Together with the proofreading mechanism, mismatch repair reduces the error rate of DNA replication to around one incorrect base pair in a billion.
Figure 3. In mismatch repair, the incorrectly added base is detected after replication. The mismatch repair proteins detect this base and remove it from the newly synthesized strand by nuclease action. The gap is now filled with the correctly paired base.
Consequences of errors in replication
Something key to think about:
Cells have evolved a variety of ways to make sure DNA errors are both detected and corrected. We have already discussed several of them. But why did so many mechanisms evolve? From proofreading by the various DNA-dependent DNA polymerases, to the complex repair systems. Such mechanisms did not evolve for errors in transcription or translation. If you are familiar with the processes of transcription and/or translation, think about what the consequences would be of an error in transcription. Would such an error affect the offspring? Would it be lethal to the cell? What about errors in translation? Ask the same questions about the process of translation. What would happen if the wrong amino acid is accidentally put into the growing polypeptide during translation? How do these contrast with DNA replication? If you are not familiar with transcription or translation, don't fret. We'll learn those soon and return to this question again.
Other DNA repair mechanisms for damaged bases
Besides errors of replication, various types of DNA damage takes place after replication. Such uncorrected errors or DNA damage may lead to serious consequences. Therefore, Nature has evolved several mechanisms for repairing damaged or incorrectly synthesized DNA.
Damaged or inappropriate bases can be repaired by several mechanisms:
- Direct chemical reversal of the damage
- Excision Repair, in which the damaged base or bases are removed and then replaced with the correct ones in a localized burst of DNA synthesis. There are three modes of excision repair, each of which employs specialized sets of enzymes.
- Base Excision Repair (BER)
- Nucleotide Excision Repair (NER)
- Mismatch Repair (MMR) (see above)
Direct Reversal of Base Damage
Direct reversal of base damage is the simplest form of DNA repair and also, the most energy efficient method. It does not require a reference template unlike the other single-strand repair mechanism. Moreover, it does not involve the process of breaking the phosphodiester backbone of the DNA. Perhaps the most frequent cause of point mutations in humans is the spontaneous addition of a methyl group (CH3-) (an example of alkylation) to Cs followed by deamination to a T. Most of these changes are repaired by enzymes, called DNA glycosylases, that remove the mismatched T restoring the correct C. This is done without the need to break the DNA backbone (in contrast to the mechanisms of excision repair described below).
Base Excision Repair (BER)
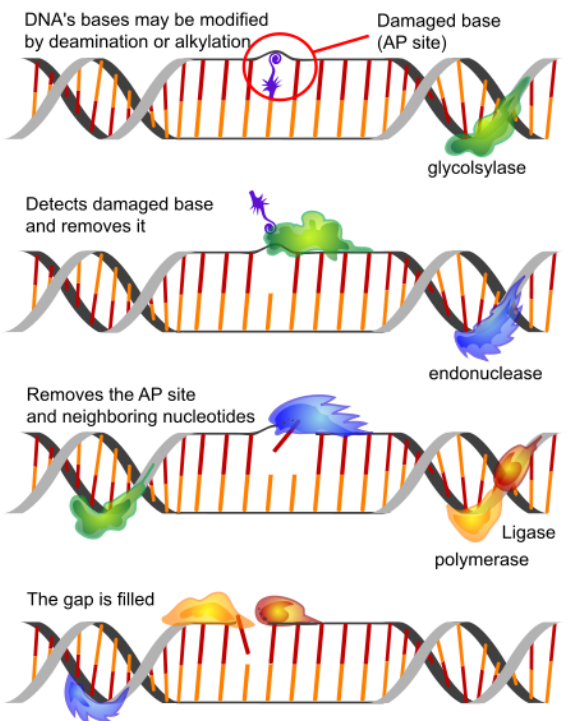
Base excision repair (BER) removes the damaged base (estimated to occur some 20,000 times a day in each cell in our body!) such as uracil by a DNA glycosylase. Cytosines in DNA sometimes undergo deamination to form the base uracil. Because cytosines pair with guanines and uracils pair with adenine, the conversion of cytosine to uracil in the DNA would lead to the insertion of an A in the newly replicated strand instead of the G that should have gone in across from a C. To prevent this from happening, uracils are removed from DNA by base excision repair. In base excision repair, a single base is first removed from the DNA, followed by removal of a region of the DNA surrounding the missing base. The gap is then repaired.
 5.2.6
5.2.6
Figure 4. Base excision repairs.
Nucleotide Excision Repair (NER)
Nucleotide excision repair enzymes replace incorrect bases by making a cut on both the 3' and 5' ends of the incorrect base. The entire segment of DNA is removed and replaced with correctly paired nucleotides by the action of a DNA polymerase. Once the bases are filled in, it seal the remaining gap with a phosphodiester linkage catalyzed by the enzyme DNA ligase. This repair mechanism is often employed when UV exposure causes the formation of pyrimidine dimers.
Figure 4. Nucleotide excision repairs thymine dimers. When exposed to UV, thymines lying adjacent to each other can form thymine dimers. In normal cells, they are excised and replaced.
Repairing Strand Breaks
Ionizing radiation and certain chemicals can produce both single-strand breaks (SSBs) and double-strand breaks (DSBs) in the DNA backbone.
Single-Strand Breaks (SSBs)
Breaks in a single strand of the DNA molecule are repaired using the same enzyme systems that are used in Base-Excision Repair (BER).
Double-Strand Breaks (DSBs)
There are two mechanisms by which the cell attempts to repair a complete break in a DNA molecule (we will cover these mechanisms in a later lecture):
- Direct joining of the broken ends. This requires proteins that recognize and bind to the exposed ends and bring them together for ligating. They would prefer to see some complementary nucleotides but can proceed without them so this type of joining is also called Nonhomologous End-Joining (NHEJ). Errors in direct joining may be a cause of the various translocations that are associated with cancers. Some examples include Burkitt's lymphoma, the Philadelphia chromosome in chronic myelogenous leukemia (CML) and B-cell leukemia.
- Homologous Recombination (also known as Homology-Directed Repair — HDR). Here the broken ends are repaired using the information on the intact sister chromatid (available in G2 after chromosome duplication), or on the homologous chromosome (in G1; that is, before each chromosome has been duplicated). This requires searching around in the nucleus for the homolog — a task sufficiently uncertain that G1 cells usually prefer to mend their DSBs by NHEJ. or on the same chromosome if there are duplicate copies of the gene on the chromosome oriented in opposite directions (head-to-head or back-to-back).
Link to external resources
What are telomeres? | Telomere animation
https://www.youtube.com/watch?v=U0fRAr-ZHCo
Telomere replication
https://www.youtube.com/watch?v=2NS0jBPurWQ
DNA repair
https://www.youtube.com/watch?v=vP8-5Bhd2ag