3.1: Origins of Mutations
- Page ID
- 25741
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Learning Objectives
- Identify some causes and types of mutations.
- Define mutagen and predict the effects of mutagens on DNA sequences.
Introduction to mutations
Mutations have a bad reputation, BUT they are ESSENTIAL to life! Watch the video for an introduction to why understanding mutations is so important for us.
Video \(\PageIndex{1}\): Introduction to mutations. (CC-BY Leacock https://www.youtube.com/watch?v=-BVFRB2hsCI&t=3s)
Types of mutations
Mutations may involve the loss (deletion), gain (insertion) of one or more base pairs, or else the substitution of one or more base pairs with another DNA sequence of equal length. These changes in DNA sequence can arise in many ways, some of which are spontaneous and due to natural processes, while others are induced by humans intentionally (or unintentionally) using mutagens. There are many ways to classify mutagens, which are the agents or processes that cause mutation or increase the frequency of mutations. We will classify mutagens here as being (1) biological, (2) chemical, or (3) physical.
Mutations of biological origin
A major source of spontaneous mutation is errors that arise during DNA replication. DNA polymerases are usually very accurate in adding a base to the growing strand that is the exact complement of the base on the template strand. However, occasionally, an incorrect base is inserted. Usually, the DNA polymerase proofreading activity will recognize and repair mispaired bases, but nevertheless, some errors become permanently incorporated in a daughter strand, and so become mutations that will be inherited by the cell’s descendents.

Figure \(\PageIndex{1}\): Mispairing of bases (e.g. G with T) can occur due to tautomerism, alkylating agents, or other effects. As a result, in this example the AT base pair in the original DNA strand will become permanently substituted by a GC based pair in some progeny. The mispaired GT pair will likely be repaired or eliminated before further rounds of replication. (Original-Deyholos-CC:AN)
The most common insult to the DNA of living organisms is depurination, in which the bond between an adenine or guanine and the deoxyribose is hydrolyzed. Depyrimidination of cytosine and thymine residues can also occur, but do so at a much slower rate than depurination. Despite the high rate of loss of these bases, they are generally identified and remediated by base excision repair (BER). Therefore, it is rare for depurination or depyrimidination to lead to mutation.
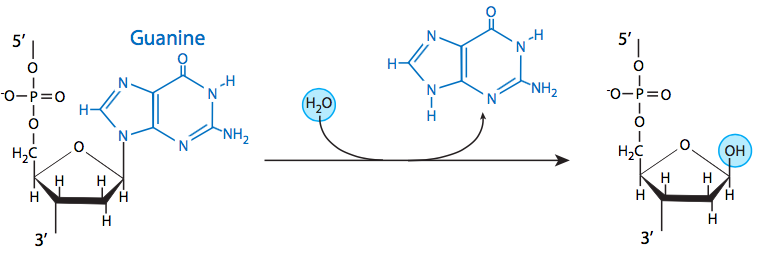
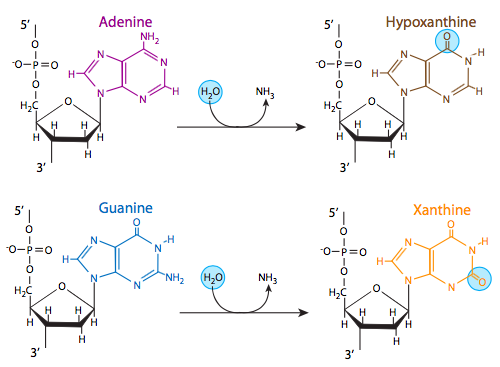
Three of the four DNA bases, adenine, guanine, and cytosine, contain amine (-NH2) groups that can be lost in a variety of pH and temperature-dependent reactions that convert the bases to hypoxanthine, xanthine, and uracil, respectively. This deamination can sometimes lead to permanent mutations during replication. Hypoxanthine base pairs with C (cytosine); xanthine base pairs with cytosine; uracil base pairs with adenine. If hypoxanthine is present when DNA polymerase is replicating DNA, a C would be incorporated instead of T; similarly if uracil is present an A would be incorporated instead of G.
Query \(\PageIndex{1}\)
Another deamination, of the modified base methylcytosine, can also lead to a mutation upon replication. Some cytosines may be methylated as part of a regulatory process to inactivate certain genes in eukaryotes, or in prokaryotes as protection against restriction endonucleases. When the methylated cytosine is deaminated, it produces a thymine, which changes the complementary nucleotide (upon replication) from a guanine to an adenine. Deamination of cytosines occurs at nearly the same rate as depurination, but deamination of other bases are not as pervasive: deamination of adenines, for example, is 50 times less likely than deamination of cytosine.

Query \(\PageIndex{2}\)
Another type of error introduced during replication is caused by a rare, temporary misalignment of a few bases between the template strand and daughter strand (Figure \(\PageIndex{5}\)). This strand-slippage causes one or more bases on either strand to be temporarily displaced in a loop that is not paired with the opposite strand. If this loop forms on the template strand, the bases in the loop may not be replicated, and a deletion will be introduced in the growing daughter strand. Conversely, if a region of the daughter strand that has just been replicated becomes displaced in a loop, this region may be replicated again, leading to an insertion of additional sequence in the daughter strand, as compared to the template strand.

Regions of DNA that have several repeats of the same few nucleotides in a row are especially prone to this type of error during replication. Thus regions with short tanderm repeats (STRs or short sequence repeats SSRs) are tend to be highly polymorphic, and are therefore particularly useful in genetics. They are also called microsatellites.
Mutations can also be caused by the insertion of viruses, transposable elements (transposons), see below, and other types of DNA that are naturally added at more or less random positions in chromosomes. The insertion may disrupt the coding or regulatory sequence of a gene, including the fusion of part of one gene with another. These insertions can occur spontaneously, or they may also be intentionally stimulated in the laboratory as a method of mutagenesis called transposon-tagging. For example, a type of transposable element called a P element is widely used in Drosophila as a biological mutagen. T-DNA, which is an insertional element modified from a bacterial pathogen, is used as a mutagen in some plant species.
Mutations due to Transposable Elements
Transposable elements (TEs) are also known as mobile genetic elements, or more informally as jumping genes. They are present throughout the chromosomes of almost all organisms. These DNA sequences have a unique ability to be cut or copied from their original location and inserted into new locations in the genome. This is called transposition. These insert locations are not entirely random, but TEs can, in principle, be inserted into almost any region of the genome. TEs can therefore insert into genes, disrupting its function and causing a mutation. Researchers have developed methods of artificially increasing the rate of transposition, which makes some TEs a useful type of mutagen. However, the biological importance of TEs extends far beyond their use in mutant screening. TEs are also important causes of disease and phenotypic instability, and they are a major mutational force in evolution.
There are two major classes of TEs in eukaryotes:
- Class I elements include retrotransposons; these transpose by means of an RNA intermediate. The TE transcript is reverse transcribed into DNA before being inserted elsewhere in the genome through the action of enzymes such as integrase.
- Class II elements are known also as transposons. They do not use reverse transcriptase or an RNA intermediate for transposition. Instead, they use an enzyme called transposase to cut DNA from the original location and then this excised dsDNA fragment is inserted into a new location.

TEs are relatively short DNA sequences (100-10,000 bp), and encode no more than a few proteins (if any). Normally, the protein-coding genes within a TE are all related to the TE’s own transposition functions. These proteins may include reverse transcriptase, transposase, and integrase. However, some TEs (of either Class I or II) do not encode any proteins at all. These non-autonomous TEs can only transpose if they are supplied with enzymes produced by other, autonomous TEs located elsewhere in the genome. In all cases, enzymes for transposition recognize conserved nucleotide sequences within the TE, which dictate where the enzymes begin cutting or copying.
The human genome consists of nearly 45% TEs, the vast majority of which are families of Class I elements called LINEs (long intersperse nuclear elements) and SINEs (short interspersed nuclear elements). The short, Alu type of SINE occurs in more than one million copies in the human genome (compare this to the approximately 21,000, non-TE, protein-coding genes in humans). Indeed, TEs make up a significant portion of the genomes of almost all eukaryotes. Class I elements, which usually transpose via an RNA copy-and-paste mechanism, tend to be more abundant than Class II elements, which mostly use a cut-and-paste mechanism. But even the cut-paste mechanism can lead to an increase in TE copy number. For example, if the site vacated by an excised transposon is repaired with a DNA template from a homologous chromosome that itself contains a copy of a transposon, then the total number of transposons in the genome will increase.
Besides greatly expanding the overall DNA content of genomes, TEs contribute to genome evolution in many other ways. As already mentioned, they may disrupt gene function by insertion into a gene’s coding region or regulatory region. More interestingly adjacent regions of chromosomal DNA are sometimes mistakenly transposed along with the TE; this can lead to gene duplication. The duplicated genes are then free to evolve independently, leading in some cases to the development of new functions. The breakage of strands by TE excision and integration can disrupt genes, and can lead to chromosome rearrangement or deletion if errors are made during strand rejoining. Furthermore, having so many similar TE sequences distributed throughout a chromosome sometimes allows mispairing of regions of homologous chromosomes at meiosis, which can cause unequal crossing-over, resulting in deletion or duplication of large segments of chromosomes. Thus, TEs are a potentially important evolutionary force, and may not be included as merely “junk DNA”, as they once were.
Video \(\PageIndex{2}\): Jumping Gene Caught in the Act (dnalc.org via https://www.youtube.com/watch?v=9MPiRx3SPMM)
Are my genes always jumping?!
If the human genome is 45% transposable elements and these elements can move around, then how does our genome stay relatively constant?
Some factors that help keep the genome (mostly) intact include:
- Many of the TE elements have acquired mutations that render them unable to transpose.
- Regulatory RNAs called piRNAs (piwi-related RNAs) appear to suppress transposition.
- In which tissue(s) would you expect piRNAs to be most active?
Mutations of Chemical Origins
Many chemical compounds, whether natural or synthetic, can react with DNA to cause mutations. In some of these reactions the chemical structure of particular bases may change, so that they are misread during replication. In others the chemical mutagens distort the double helix causing it to be replicated inaccurately, while still others may cause breaks in chromosomes that lead to deletions and other types of aberrations. The following are examples of two classes of chemical mutagens that are important in genetics and medicine: alkylating agents, and intercalating agents.
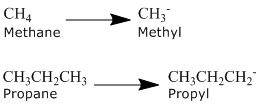
What is an alkyl group?
An alkyl group is formed by removing one hydrogen from an alkane chain (CnH2n+2) and is described by the formula CnH2n+1. The removal of this hydrogen results in a stem change from -ane to -yl. Take a look at the following examples.
Ethane methyl sulfonate (EMS) is an example of an alkylating agent that is commonly used by geneticists to induce mutations in a wide range of both prokaryotes and eukaryotes. The organism is fed or otherwise exposed to a solution of EMS. It reacts with some of the G bases it encounters in a process called alkylation, where the addition of an alkyl group to G changes the base pairing properties so that the next time the alkylated DNA strand is replicated, a T instead of a C will be inserted opposite to the alkylated-G in the daughter strand. The new strand therefore bears a mutation (base substitution), which will be inherited in all the strands that are subsequently replicated from it.

Figure \(\PageIndex{7}\): Alkylation of G (shown in red) allows G to bond with T, rather than with C. (Original-Deyholos-CC:AN)
Intercalating agents are another type of chemical mutagen. They tend to be flat, planar molecules like benzo[a]pyrene, a component of wood and tobacco smoke, and induce mutations by inserting between the stacked bases at the center of the DNA helix. This intercalation distorts the shape of the DNA helix, which can cause the wrong bases to be added to a growing DNA strand during DNA synthesis.
There are a large number of chemicals that act as intercalating agents, can mutate DNA, and are carcinogenic (can cause cancer). Many of these are also used to treat cancer, as they preferentially kill actively dividing cells. Another important intercalating agent is ethidium bromide, the fluorescent dye that stains DNA in laboratory assays. For this reason, molecular biologists are trained to handle this chemical carefully.
Exercise \(\PageIndex{1}\)
The chemical EMS adds an ethyl group to G, which then pairs with T instead of C. What would be the effect of exposing the DNA sequence ...CATGTCA... to EMS?
- Answer
-
When DNA is exposed to a mutagen it is unlikely that all the Gs in a given sequence will be affected. We will assume that only the G in the above sequence is affected (not the two Gs that will be on the complementary strand). I will write the modified sequence as ...CATGmTCA...
- When the methylated strand is used as a template for DNA replication, the newly synthesized strand will be ...GTATAGT...
- If the cell undergoes another round of cell division, using the new strand as a template will create the strand ...CATATCA... and the former GC base pair is now an AT base pair. The cell has no way of knowing that this base pair is incorrect or whether or not it will affect the cell.
Mutations of physical origin
Anything that damages DNA by transferring energy to it can be considered a physical mutagen. Usually this involves radioactive particles, x-rays, or UV light. The smaller, fast moving particles may cause base substitutions or delete a single bases, while larger, slightly slower particles may induce larger deletions by breaking the double stranded helix of the chromosome. Physical mutagens can also create unusual structures in DNA, such as the thymine dimers formed by UV light (Figure \(\PageIndex{9}\)). Thymine dimers disrupt normal base-pairing in the double helix, and may block replication altogether if not repaired by the cell’s DNA repair enzymes.
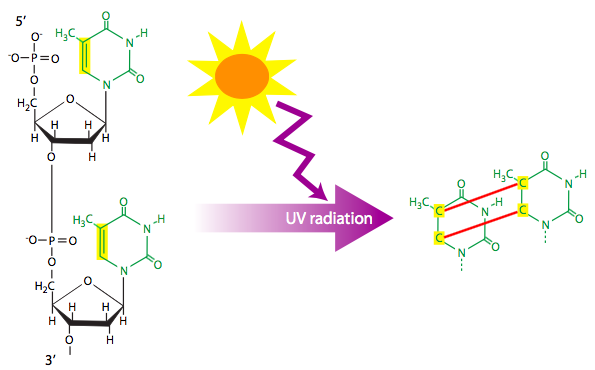
Figure \(\PageIndex{9}\): Ultraviolet radiation can be absorbed by some DNA and commonly causes pyrimidine cyclobutyl dimers connecting adjacent nucleotide bases. (CC Wong via 7.5: DNA Lesions)
Summary Table of Mutagenic Agents
| Mutagenic Agents | Mode of Action | Effect on DNA | Resulting Type of Mutation |
|---|---|---|---|
| Nucleoside analogs | |||
| 2-aminopurine | Is inserted in place of A but base pairs with C | Converts AT to GC base pair | Point |
| 5-bromouracil | Is inserted in place of T but base pairs with G | Converts AT to GC base pair | Point |
| Nucleotide-modifying agent | |||
| Nitrous oxide | Deaminates C to U | Converts GC to AT base pair | Point |
| Ethane methyl sulfonate (EMS) | Alkylates G (which pairs with T) | ||
| Intercalating agents | |||
| Acridine orange, ethidium bromide, polycyclic aromatic hydrocarbons | Distorts double helix, creates unusual spacing between nucleotides | Introduces small deletions and insertions | Frameshift |
| Ionizing radiation | |||
| X-rays, γ-rays | Forms hydroxyl radicals | Causes single- and double-strand DNA breaks | Repair mechanisms may introduce mutations |
| X-rays, γ-rays | Modifies bases (e.g., deaminating C to U) | Converts GC to AT base pair | Point |
| Nonionizing radiation | |||
| Ultraviolet | Forms pyrimidine (usually thymine) dimers | Causes DNA replication errors | Frameshift or point |
Query \(\PageIndex{3}\)
Contributors and Attributions
Dr. Todd Nickle and Isabelle Barrette-Ng (Mount Royal University) The content on this page is licensed under CC SA 3.0 licensing guidelines.
- E. V. Wong Axolotl Academica Publishing (Biology) at Axolotl Academica Publishing
- Mutations. (2021, January 3). Retrieved August 6, 2021, from https://bio.libretexts.org/@go/page/5184 " Mutations" by OpenStax, LibreTexts is licensed under CC BY (11.5: Mutations).


