9: Transformation of ligated plasmid DNA
- Page ID
- 169773
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Introduction:
Transformation is the idea of transferring DNA from one organism to another. This is generally done by breaking done the membrane of the cell, allowing DNA to move into the cell. Then the cell membrane is reconstructed and the new DNA is now part of the cell. In this experiment, pGFPuv (ligated circular and unligated lineal, and a 1ng control plasmid (pGFPuv) used for measuring transformation efficiency) will be used for transformation.
Transformation & Storage Solution (2X TSS) enables researchers to take advantage of the simple system described by Chung et al.1 for the preparation, long-term storage and transformation of competent E. coli. Early log-phase cells are suspended in 1X TSS: a solution containing polyethylene glycol, dimethyl sulfoxide, and divalent cations in a bacterial growth medium. The transforming DNA is added to the TSS-treated cells, the mixture is incubated on ice, and the transformed cells are plated on selective media. Transformation efficiencies of E6-E8 colonies/ug of DNA are routinely obtained without lengthy host cell preparation or heat shock (1, 2). For example, E. coli strains DH1, DH5, HB101, JM109, LE392, MM294, SCS-1, and XL-1 gave transformation efficiencies of 1.5-6 x E7 colonies/ug of DNA.
Precise transformation efficiencies will depend on both the host strain and the nature and quality of the transforming DNA. For added convenience, TSS-treated competent cells can be prepared in advance and stored at –70C for later use with little or no loss in transformation efficiency. For example, TSS-treated competent E. coli JM109 cells showed no significant loss of transformation efficiency after 1-18 weeks at –70C. However, the transformation efficiency of TSS-treated E. coli K802 competent cells was reduced several-fold after similar treatment. Fresh TSS-treated competent cells should be used when the total number of recombinants, the ligation or transformation efficiency or the percentage of correct recombinants is expected to be low.
References :
1. Chung, C.T. et al., (1989) Proc. Natl. Acad. Sci. USA 86, 2172.
2. Groth, D. et al., (1979) Anal. Biochem. 240, 302.
One-Step Preparation of Competent E. coli Cells by TSS
1. Grow cells in 3 ml LB at 37C to early exponential phase (OD600=0.3-0.4).(ex. make a 1/50 dilution (60 ul) of a overnight culture into a fresh LB medium and grow for about 1 hr)
2. Make (1ml aliquots cells in autoclaved 1.5ml tubes)*3.
3. Pellet cells by centrifugation at 1000 xg for 10 min. at 4ºC (or 12,000 rpm for 1 min. in a mini-centrifuge).
4. Resuspend cells of each tube in 1/20 their original volume (50ul) in ice-cold TSS solution.
5. For storage, freeze immediately in liquid nitrogen (or dry ice in ethanol) and store at -70 ºC.
6. Add 2.5 ul of ligated, unligated plasmid DNA and 1ul of control plasmid (see the following Table) to the competent cells (corresponding to 100 pg-10 ng of tansforming DNA)
7. Mix the cells and DNA and place on ice for 10 min, then at room temperature for 10 min, and then again on ice for 10 min.
8. Add 0.5 ml of LB broth and incubate the tubes at 37ºC for 1hr.
9. A brief spin and remove 450 ul of the supernatant.
10. Resuspend the cell ppt and plate the cells on the appropriate selective media.
Table. Transformation set up
|
1 |
2 |
3 |
|
|
Transformation |
ligation |
Negative control (no ligase) |
uncut pGFPuv control (1 ng/ul) |
|
DNA |
2.5 ul |
2.5 ul |
2.5 ul |
|
Competent cells |
50 ul |
50 ul |
50 ul |
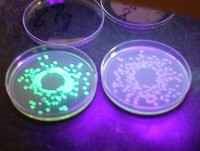
Figure. Fluorescent phenotype of a petri-dish with ampicillin on a UV lamp. DH5a/pGFPuv clone (Left) and a fluorescent negative control clone (Right).
Table. Preparation of transformation and storage solution (TSS)
|
LB |
For 100 mL LB, add the following: |
|
10% PEG 8000 |
10 g PEG 8000 |
|
20 mM glucose |
1 ml of 2M glucose |
|
5% DMSO |
5 mL DMSO |
|
10 mM MgCl2 |
1 ml of 1M MgCl2 |
|
10 mM MgSO4 |
1 ml of 1M MgSO4 |
Note: Final pH should be 6.5
|
|
|
Fig. Scheme for the transformation



