6.3: Isolation, Culture, and Identification of Viruses
- Page ID
- 75844
- Discuss why viruses were originally described as filterable agents
- Describe the cultivation of viruses and specimen collection and handling
- Compare in vivo and in vitro techniques used to cultivate viruses
At the beginning of this chapter, we described how porcelain Chamberland filters with pores small enough to allow viruses to pass through were used to discover TMV. Today, porcelain filters have been replaced with membrane filters and other devices used to isolate and identify viruses.
Isolation of Viruses
Unlike bacteria, many of which can be grown on an artificial nutrient medium, viruses require a living host cell for replication. Infected host cells (eukaryotic or prokaryotic) can be cultured and grown, and then the growth medium can be harvested as a source of virus. Virions in the liquid medium can be separated from the host cells by either centrifugation or filtration. Filters can physically remove anything present in the solution that is larger than the virions; the viruses can then be collected in the filtrate (Figure \(\PageIndex{1}\)).
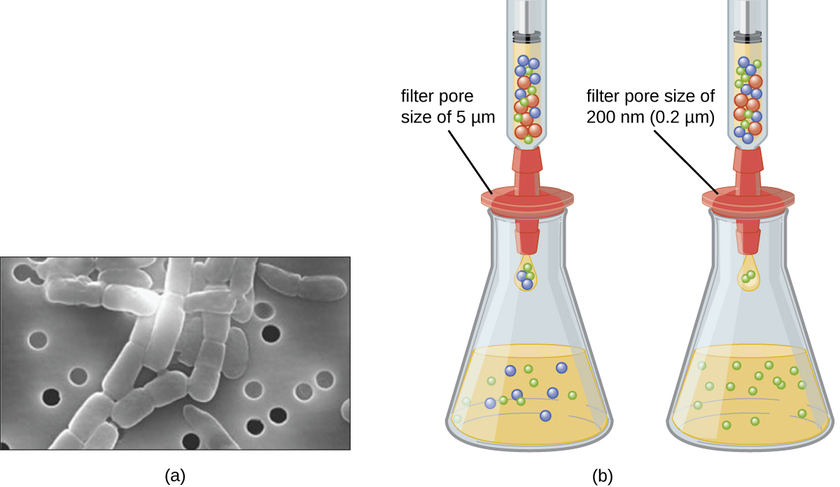
What size filter pore is needed to collect a virus?
Cultivation of Viruses
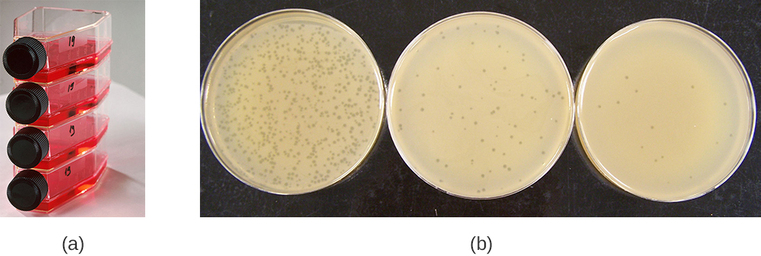
Viruses can be grown in vivo (within a whole living organism, plant, or animal) or in vitro (outside a living organism in cells in an artificial environment, such as a test tube, cell culture flask, or agar plate). Bacteriophages can be grown in the presence of a dense layer of bacteria (also called a bacterial lawn) grown in a 0.7 % soft agar in a Petri dish or flat (horizontal) flask (Figure \(\PageIndex{2a}\)). The agar concentration is decreased from the 1.5% usually used in culturing bacteria. The soft 0.7% agar allows the bacteriophages to easily diffuse through the medium. For lytic bacteriophages, lysing of the bacterial hosts can then be readily observed when a clear zone called a plaque is detected (Figure \(\PageIndex{1b}\)). As the phage kills the bacteria, many plaques are observed among the cloudy bacterial lawn.
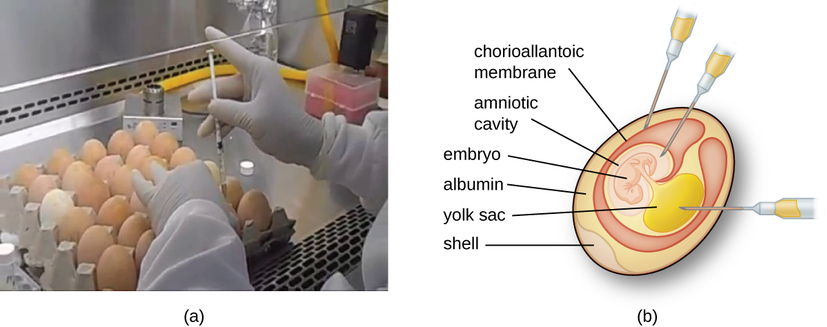
Animal viruses require cells within a host animal or tissue-culture cells derived from an animal. Animal virus cultivation is important for 1) identification and diagnosis of pathogenic viruses in clinical specimens, 2) production of vaccines, and 3) basic research studies. In vivo host sources can be a developing embryo in an embryonated bird’s egg (e.g., chicken, turkey) or a whole animal. For example, most of the influenza vaccine manufactured for annual flu vaccination programs is cultured in hens’ eggs.
The embryo or host animal serves as an incubator for viral replication (Figure \(\PageIndex{3}\)). Location within the embryo or host animal is important. Many viruses have a tissue tropism, and must therefore be introduced into a specific site for growth. Within an embryo, target sites include the amniotic cavity, the chorioallantoic membrane, or the yolk sac. Viral infection may damage tissue membranes, producing lesions called pox; disrupt embryonic development; or cause the death of the embryo.
For in vitro studies, various types of cells can be used to support the growth of viruses. A primary cell culture is freshly prepared from animal organs or tissues. Cells are extracted from tissues by mechanical scraping or mincing to release cells or by an enzymatic method using trypsin or collagenase to break up tissue and release single cells into suspension. Because of anchorage-dependence requirements, primary cell cultures require a liquid culture medium in a Petri dish or tissue-culture flask so cells have a solid surface such as glass or plastic for attachment and growth. Primary cultures usually have a limited life span. When cells in a primary culture undergo mitosis and a sufficient density of cells is produced, cells come in contact with other cells. When this cell-to-cell-contact occurs, mitosis is triggered to stop. This is called contact inhibition and it prevents the density of the cells from becoming too high. To prevent contact inhibition, cells from the primary cell culture must be transferred to another vessel with fresh growth medium. This is called a secondary cell culture. Periodically, cell density must be reduced by pouring off some cells and adding fresh medium to provide space and nutrients to maintain cell growth. In contrast to primary cell cultures, continuous cell lines, usually derived from transformed cells or tumors, are often able to be subcultured many times or even grown indefinitely (in which case they are called immortal). Continuous cell lines may not exhibit anchorage dependency (they will grow in suspension) and may have lost their contact inhibition. As a result, continuous cell lines can grow in piles or lumps resembling small tumor growths (Figure \(\PageIndex{4}\)).
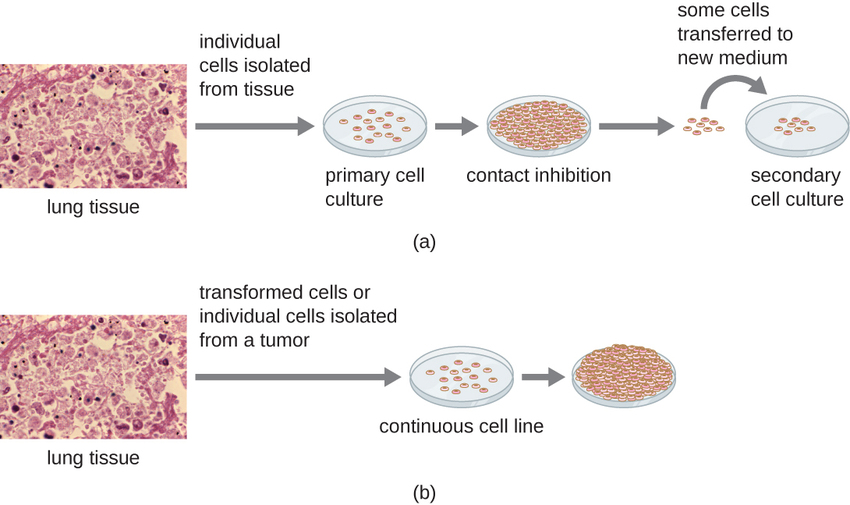
An example of an immortal cell line is the HeLa cell line, which was originally cultivated from tumor cells obtained from Henrietta Lacks, a patient who died of cervical cancer in 1951. HeLa cells were the first continuous tissue-culture cell line and were used to establish tissue culture as an important technology for research in cell biology, virology, and medicine. Prior to the discovery of HeLa cells, scientists were not able to establish tissue cultures with any reliability or stability. More than six decades later, this cell line is still alive and being used for medical research. See below (The Immortal Cell Line of Henrietta Lacks) to read more about this important cell line and the controversial means by which it was obtained.
What property of cells makes periodic dilutions of primary cell cultures necessary?
In January 1951, Henrietta Lacks, a 30-year-old African American woman from Baltimore, was diagnosed with cervical cancer at John Hopkins Hospital. We now know her cancer was caused by the human papillomavirus (HPV). Cytopathic effects of the virus altered the characteristics of her cells in a process called transformation, which gives the cells the ability to divide continuously. This ability, of course, resulted in a cancerous tumor that eventually killed Mrs. Lacks in October at age 31. Before her death, samples of her cancerous cells were taken without her knowledge or permission. The samples eventually ended up in the possession of Dr. George Gey, a biomedical researcher at Johns Hopkins University. Gey was able to grow some of the cells from Lacks’s sample, creating what is known today as the immortal HeLa cell line. These cells have the ability to live and grow indefinitely and, even today, are still widely used in many areas of research.
According to Lacks’s husband, neither Henrietta nor the family gave the hospital permission to collect her tissue specimen. Indeed, the family was not aware until 20 years after Lacks’s death that her cells were still alive and actively being used for commercial and research purposes. Yet HeLa cells have been pivotal in numerous research discoveries related to polio, cancer, and AIDS, among other diseases. The cells have also been commercialized, although they have never themselves been patented. Despite this, Henrietta Lacks’s estate has never benefited from the use of the cells, although, in 2013, the Lacks family was given control over the publication of the genetic sequence of her cells.
This case raises several bioethical issues surrounding patients’ informed consent and the right to know. At the time Lacks’s tissues were taken, there were no laws or guidelines about informed consent. Does that mean she was treated fairly at the time? Certainly by today’s standards, the answer would be no. Harvesting tissue or organs from a dying patient without consent is not only considered unethical but illegal, regardless of whether such an act could save other patients’ lives. Is it ethical, then, for scientists to continue to use Lacks’s tissues for research, even though they were obtained illegally by today’s standards?
Ethical or not, Lacks’s cells are widely used today for so many applications that it is impossible to list them all. Is this a case in which the ends justify the means? Would Lacks be pleased to know about her contribution to science and the millions of people who have benefited? Would she want her family to be compensated for the commercial products that have been developed using her cells? Or would she feel violated and exploited by the researchers who took part of her body without her consent? Because she was never asked, we will never know.
Detection of a Virus
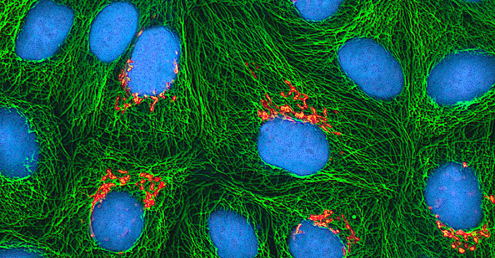
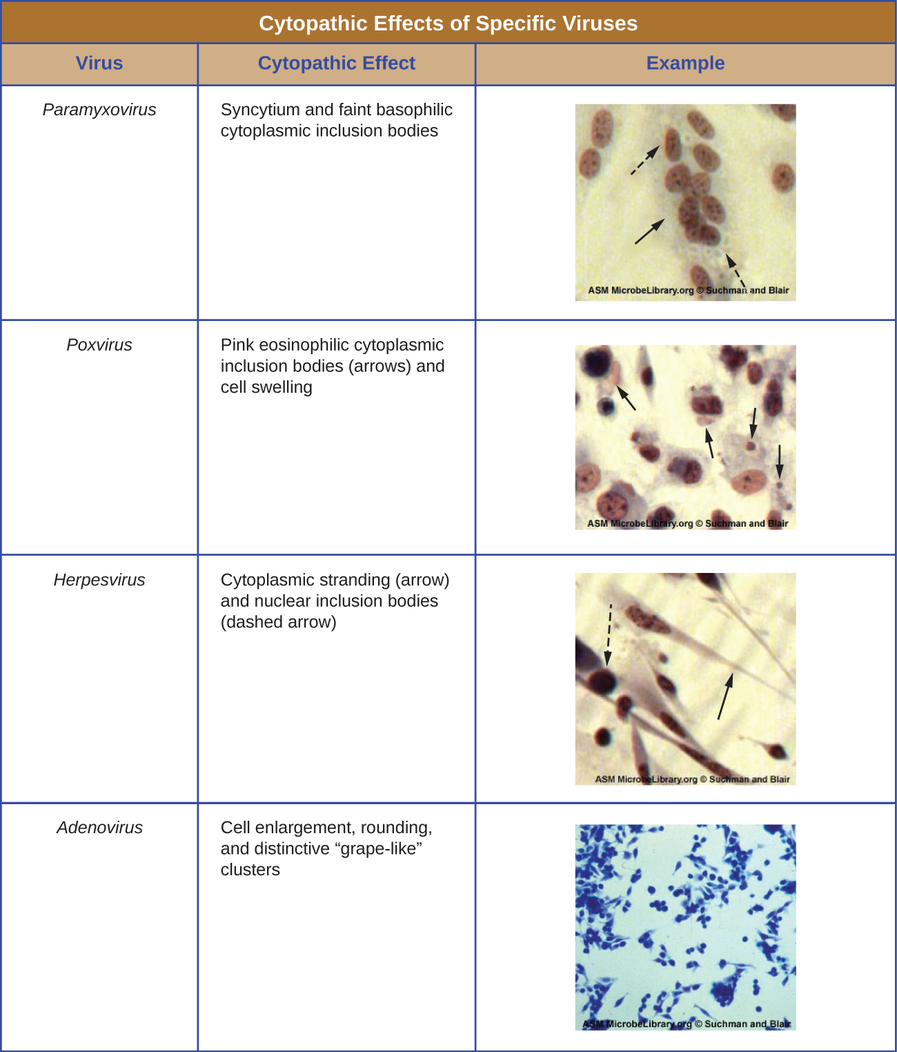
Regardless of the method of cultivation, once a virus has been introduced into a whole host organism, embryo, or tissue-culture cell, a sample can be prepared from the infected host, embryo, or cell line for further analysis under a brightfield, electron, or fluorescent microscope. Cytopathic effects (CPEs) are distinct observable cell abnormalities due to viral infection. CPEs can include loss of adherence to the surface of the container, changes in cell shape from flat to round, shrinkage of the nucleus, vacuoles in the cytoplasm, fusion of cytoplasmic membranes and the formation of multinucleated syncytia, inclusion bodies in the nucleus or cytoplasm, and complete cell lysis (see Figure \(\PageIndex{6}\)).
Further pathological changes include viral disruption of the host genome and altering normal cells into transformed cells, which are the types of cells associated with carcinomas and sarcomas. The type or severity of the CPE depends on the type of virus involved. Figure \(\PageIndex{6}\) lists CPEs for specific viruses.
Watch this video to learn about the effects of viruses on cells.
Hemagglutination Assay
A serological assay is used to detect the presence of certain types of viruses in patient serum. Serum is the straw-colored liquid fraction of blood plasma from which clotting factors have been removed. Serum can be used in a direct assay called a hemagglutination assay to detect specific types of viruses in the patient’s sample. Hemagglutination is the agglutination (clumping) together of erythrocytes (red blood cells). Many viruses produce surface proteins or spikes called hemagglutinins that can bind to receptors on the membranes of erythrocytes and cause the cells to agglutinate. Hemagglutination is observable without using the microscope, but this method does not always differentiate between infectious and noninfectious viral particles, since both can agglutinate erythrocytes.
To identify a specific pathogenic virus using hemagglutination, we must use an indirect approach. Proteins called antibodies, generated by the patient’s immune system to fight a specific virus, can be used to bind to components such as hemagglutinins that are uniquely associated with specific types of viruses. The binding of the antibodies with the hemagglutinins found on the virus subsequently prevent erythrocytes from directly interacting with the virus. So when erythrocytes are added to the antibody-coated viruses, there is no appearance of agglutination; agglutination has been inhibited. We call these types of indirect assays for virus-specific antibodies hemagglutination inhibition (HAI) assays. HAI can be used to detect the presence of antibodies specific to many types of viruses that may be causing or have caused an infection in a patient even months or years after infection (see Figure \(\PageIndex{7}\)).
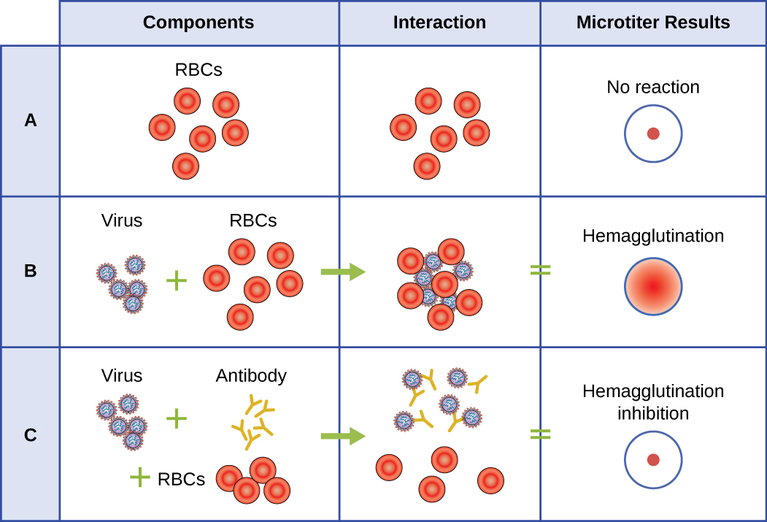
What is the outcome of a positive HIA test?
Nucleic Acid Amplification Test
Nucleic acid amplification tests (NAAT) are used in molecular biology to detect unique nucleic acid sequences of viruses in patient samples. Polymerase chain reaction (PCR) is an NAAT used to detect the presence of viral DNA in a patient’s tissue or body fluid sample. PCR is a technique that amplifies (i.e., synthesizes many copies) of a viral DNA segment of interest. Using PCR, short nucleotide sequences called primers bind to specific sequences of viral DNA, enabling identification of the virus.
Reverse transcriptase-PCR (RT-PCR) is an NAAT used to detect the presence of RNA viruses. RT-PCR differs from PCR in that the enzyme reverse transcriptase (RT) is used to make a cDNA from the small amount of viral RNA in the specimen. The cDNA can then be amplified by PCR. Both PCR and RT-PCR are used to detect and confirm the presence of the viral nucleic acid in patient specimens.
Michelle, a 21-year-old nursing student, came to the university clinic worried that she might have been exposed to a sexually transmitted disease (STD). Her sexual partner had recently developed several bumps on the base of his penis. He had put off going to the doctor, but Michelle suspects they are genital warts caused by HPV. She is especially concerned because she knows that HPV not only causes warts but is a prominent cause of cervical cancer. She and her partner always use condoms for contraception, but she is not confident that this precaution will protect her from HPV.
Michelle’s physician finds no physical signs of genital warts or any other STDs, but recommends that Michelle get a Pap smear along with an HPV test. The Pap smear will screen for abnormal cervical cells and the CPEs associated with HPV; the HPV test will test for the presence of the virus. If both tests are negative, Michelle can be more assured that she most likely has not become infected with HPV. However, her doctor suggests it might be wise for Michelle to get vaccinated against HPV to protect herself from possible future exposure.
Why does Michelle’s physician order two different tests instead of relying on one or the other?
Enzyme Immunoassay
Enzyme immunoassays (EIAs) rely on the ability of antibodies to detect and attach to specific biomolecules called antigens. The detecting antibody attaches to the target antigen with a high degree of specificity in what might be a complex mixture of biomolecules. Also included in this type of assay is a colorless enzyme attached to the detecting antibody. The enzyme acts as a tag on the detecting antibody and can interact with a colorless substrate, leading to the production of a colored end product. EIAs often rely on layers of antibodies to capture and react with antigens, all of which are attached to a membrane filter (see Figure \(\PageIndex{8}\)). EIAs for viral antigens are often used as preliminary screening tests. If the results are positive, further confirmation will require tests with even greater sensitivity, such as a western blot or an NAAT.
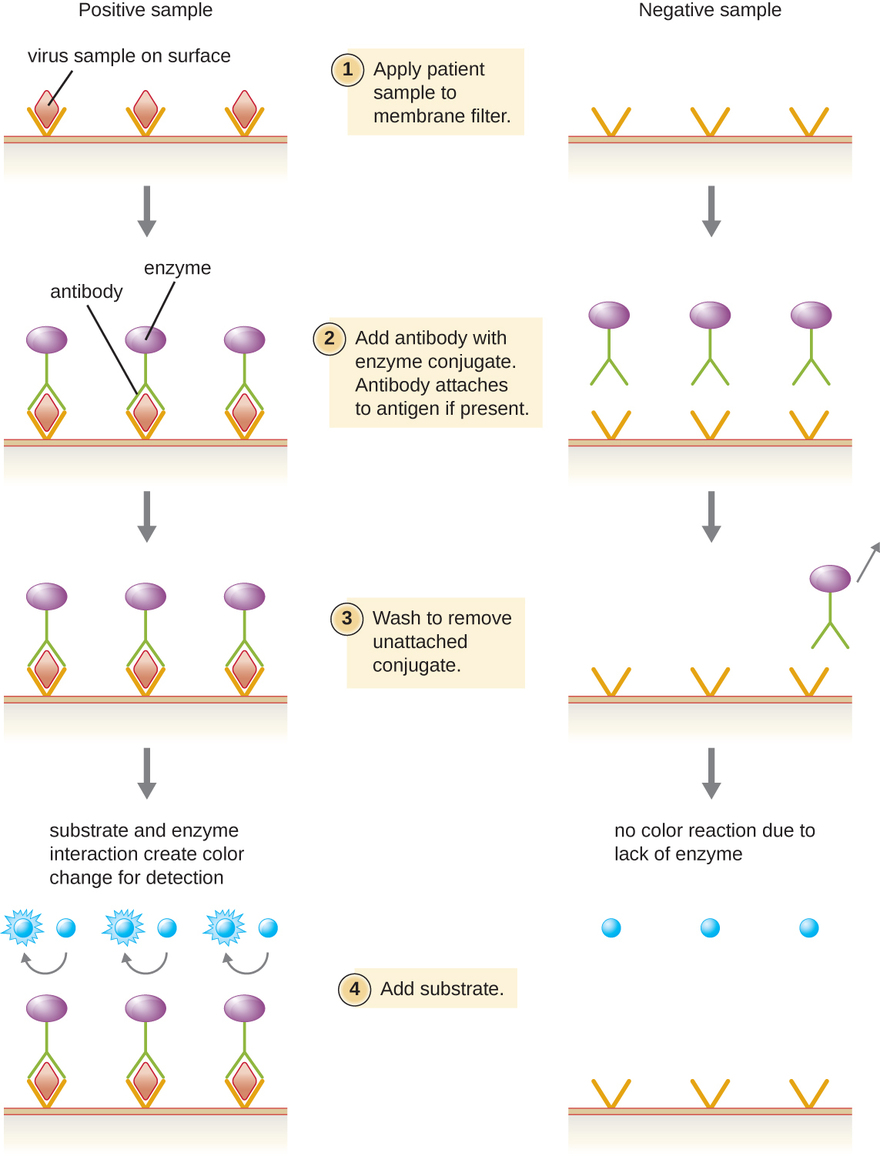
What typically indicates a positive EIA test?
Along with the RT/PCR analysis, David’s saliva was also collected for viral cultivation. In general, no single diagnostic test is sufficient for antemortem diagnosis, since the results will depend on the sensitivity of the assay, the quantity of virions present at the time of testing, and the timing of the assay, since release of virions in the saliva can vary. As it turns out, the result was negative for viral cultivation from the saliva. This is not surprising to David’s doctor, because one negative result is not an absolute indication of the absence of infection. It may be that the number of virions in the saliva is low at the time of sampling. It is not unusual to repeat the test at intervals to enhance the chance of detecting higher virus loads.
Should David’s doctor modify his course of treatment based on these test results?
Summary
- Viral cultivation requires the presence of some form of host cell (whole organism, embryo, or cell culture).
- Viruses can be isolated from samples by filtration.
- Viral filtrate is a rich source of released virions.
- Bacteriophages are detected by presence of clear plaques on bacterial lawn.
- Animal and plant viruses are detected by cytopathic effects, molecular techniques (PCR, RT-PCR), enzyme immunoassays, and serological assays (hemagglutination assay, hemagglutination inhibition assay).
Contributor
Nina Parker, (Shenandoah University), Mark Schneegurt (Wichita State University), Anh-Hue Thi Tu (Georgia Southwestern State University), Philip Lister (Central New Mexico Community College), and Brian M. Forster (Saint Joseph’s University) with many contributing authors. Original content via Openstax (CC BY 4.0; Access for free at https://openstax.org/books/microbiology/pages/1-introduction)


