13.5: Cloning
- Page ID
- 44495
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Recognize technologies used for molecular, cellular, and reproductive cloning
Molecular Cloning
In general, the word “cloning” means the creation of a perfect replica; however, in biology, the re-creation of a whole organism is referred to as “reproductive cloning.” Long before attempts were made to clone an entire organism, researchers learned how to reproduce desired regions or fragments of the genome, a process that is referred to as molecular cloning.
Cloning small fragments of the genome allows for the manipulation and study of specific genes (and their protein products), or noncoding regions in isolation. A plasmid (also called a vector) is a small circular DNA molecule that replicates independently of the chromosomal DNA of microorganisms such as E. coli. In cloning, the plasmid molecules can be used to provide a “folder” in which to insert a desired DNA fragment. Plasmids are usually introduced into a bacterial host for proliferation. In the bacterial context, the fragment of DNA from the human genome (or the genome of another organism that is being studied) is referred to as foreign DNA, or a transgene, to differentiate it from the DNA of the bacterium, which is called the host DNA.
Plasmids occur naturally in bacterial populations (such as Escherichia coli) and have genes that can contribute favorable traits to the organism, such as antibiotic resistance (the ability to be unaffected by antibiotics). Plasmids have been repurposed and engineered as vectors for molecular cloning and the large-scale production of important reagents, such as insulin and human growth hormone. An important feature of plasmid vectors is the ease with which a foreign DNA fragment can be introduced via the multiple cloning site (MCS). The MCS is a short DNA sequence containing multiple sites that can be cut with different commonly available restriction endonucleases. Restriction endonucleases recognize specific DNA sequences and cut them in a predictable manner; they are naturally produced by bacteria as a defense mechanism against foreign DNA. Many restriction endonucleases make staggered cuts in the two strands of DNA, such that the cut ends have a 2- or 4-base single-stranded overhang. Because these overhangs are capable of annealing with complementary overhangs, these are called “sticky ends.” Addition of an enzyme called DNA ligase permanently joins the DNA fragments via phosphodiester bonds. In this way, any DNA fragment generated by restriction endonuclease cleavage can be spliced between the two ends of a plasmid DNA that has been cut with the same restriction endonuclease (Figure 1).
Recombinant DNA Molecules
Plasmids with foreign DNA inserted into them are called recombinant DNA molecules because they are created artificially and do not occur in nature. They are also called chimeric molecules because the origin of different parts of the molecules can be traced back to different species of biological organisms or even to chemical synthesis. Proteins that are expressed from recombinant DNA molecules are called recombinant proteins. Not all recombinant plasmids are capable of expressing genes. The recombinant DNA may need to be moved into a different vector (or host) that is better designed for gene expression. Plasmids may also be engineered to express proteins only when stimulated by certain environmental factors, so that scientists can control the expression of the recombinant proteins.
View an animation of recombination in cloning from the DNA Learning Center.
You are working in a molecular biology lab and, unbeknownst to you, your lab partner left the foreign genomic DNA that you are planning to clone on the lab bench overnight instead of storing it in the freezer. As a result, it was degraded by nucleases, but still used in the experiment. The plasmid, on the other hand, is fine. What results would you expect from your molecular cloning experiment?
- There will be no colonies on the bacterial plate.
- There will be blue colonies only.
- There will be blue and white colonies.
- The will be white colonies only.
[reveal-answer q=”511969″]Show Answer[/reveal-answer]
[hidden-answer a=”511969″]Answer b. The experiment would result in blue colonies only.[/hidden-answer]
Cellular Cloning
Unicellular organisms, such as bacteria and yeast, naturally produce clones of themselves when they replicate asexually by binary fission; this is known as cellular cloning. The nuclear DNA duplicates by the process of mitosis, which creates an exact replica of the genetic material. Cellular cloning is often used as a tool in molecular biology studies, when an asexually reproducing organism is “cloned” in order to increase a portion of DNA added to the cell.

Reproductive Cloning
Reproductive cloning is a method used to make a clone or an identical copy of an entire multicellular organism. Most multicellular organisms undergo reproduction by sexual means, which involves genetic hybridization of two individuals (parents), making it impossible for generation of an identical copy or a clone of either parent. Recent advances in biotechnology have made it possible to artificially induce asexual reproduction of mammals in the laboratory.
Parthenogenesis, or “virgin birth,” occurs when an embryo grows and develops without the fertilization of the egg occurring; this is a form of asexual reproduction. An example of parthenogenesis occurs in species in which the female lays an egg and if the egg is fertilized, it is a diploid egg and the individual develops into a female; if the egg is not fertilized, it remains a haploid egg and develops into a male. The unfertilized egg is called a parthenogenic, or virgin, egg. Some insects and reptiles lay parthenogenic eggs that can develop into adults.
Sexual reproduction requires two cells; when the haploid egg and sperm cells fuse, a diploid zygote results. The zygote nucleus contains the genetic information to produce a new individual. However, early embryonic development requires the cytoplasmic material contained in the egg cell. This idea forms the basis for reproductive cloning. Therefore, if the haploid nucleus of an egg cell is replaced with a diploid nucleus from the cell of any individual of the same species (called a donor), it will become a zygote that is genetically identical to the donor. Somatic cell nuclear transfer is the technique of transferring a diploid nucleus into an enucleated egg. It can be used for either therapeutic cloning or reproductive cloning.
The first cloned animal was Dolly, a sheep who was born in 1996. The success rate of reproductive cloning at the time was very low. Dolly lived for seven years and died of respiratory complications (Figure 3). There is speculation that because the cell DNA belongs to an older individual, the age of the DNA may affect the life expectancy of a cloned individual. Since Dolly, several animals such as horses, bulls, and goats have been successfully cloned, although these individuals often exhibit facial, limb, and cardiac abnormalities. There have been attempts at producing cloned human embryos as sources of embryonic stem cells, sometimes referred to as cloning for therapeutic purposes. Therapeutic cloning produces stem cells to attempt to remedy detrimental diseases or defects (unlike reproductive cloning, which aims to reproduce an organism). Still, therapeutic cloning efforts have met with resistance because of bioethical considerations.
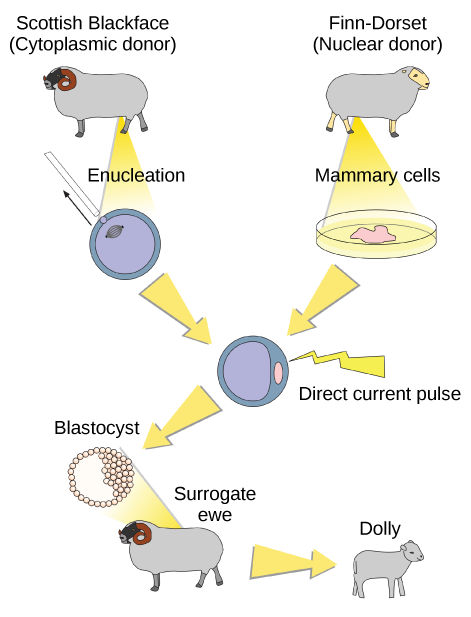
Do you think Dolly was a Finn-Dorset or a Scottish Blackface sheep?
[practice-area rows=”2″][/practice-area]
[reveal-answer q=”985505″]Show Answer[/reveal-answer]
[hidden-answer a=”985505″]Dolly was a Finn-Dorset sheep because even though the original cell came from a Scottish blackface sheep and the surrogate mother was a Scottish blackface, the DNA came from a Finn-Dorset.[/hidden-answer]
Contributors and Attributions
- Modification of DNA Cloning. Authored by: Tinojasontran. Provided by: English Wikibooks. Located at: https://commons.wikimedia.org/wiki/File:DNAcloning.jpg. License: Public Domain: No Known Copyright
- Biology. Provided by: OpenStax CNX. Located at: http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8. License: CC BY: Attribution. License Terms: Download for free at http://cnx.org/contents/185cbf87-c72...f21b5eabd@10.8

