6.6: The Prokaryotic Cell
- Page ID
- 44604
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Describe the basic structure of a typical prokaryote
All cells share four common components: (1) a plasma membrane, an outer covering that separates the cell’s interior from its surrounding environment; (2) cytoplasm, consisting of a jelly-like region within the cell in which other cellular components are found; (3) DNA, the genetic material of the cell; and (4) ribosomes, particles that synthesize proteins. Prokaryotic cells differ from eukaryotic cells in several key ways.
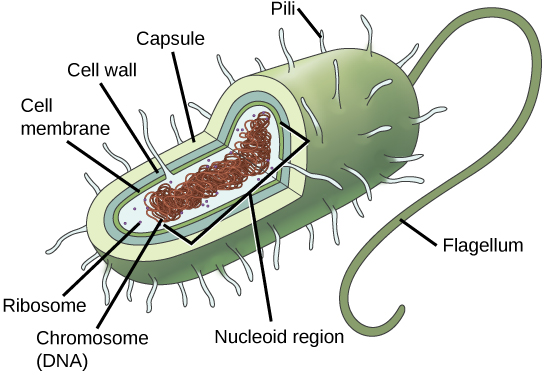
A prokaryotic cell is a simple, single-celled (unicellular) organism that lacks a nucleus, or any other membrane-bound organelle. Prokaryotic DNA is found in the central part of the cell: a darkened region called the nucleoid (Figure 1).
Some prokaryotes have flagella, pili, or fimbriae. Flagella are used for locomotion, while most pili are used to exchange genetic material during a type of reproduction called conjugation. Many prokaryotes also have a cell wall and capsule. The cell wall acts as an extra layer of protection, helps the cell maintain its shape, and prevents dehydration. The capsule enables the cell to attach to surfaces in its environment.
Reproduction
Reproduction in prokaryotes is asexual and usually takes place by binary fission. Recall that the DNA of a prokaryote exists as a single, circular chromosome. Prokaryotes do not undergo mitosis. Rather the chromosome is replicated and the two resulting copies separate from one another due to the growth of the cell. The prokaryote, now enlarged, is pinched inward at its equator and the two resulting cells, which are clones, separate. Binary fission does not provide an opportunity for genetic recombination or genetic diversity, but prokaryotes can share genes by three other mechanisms.
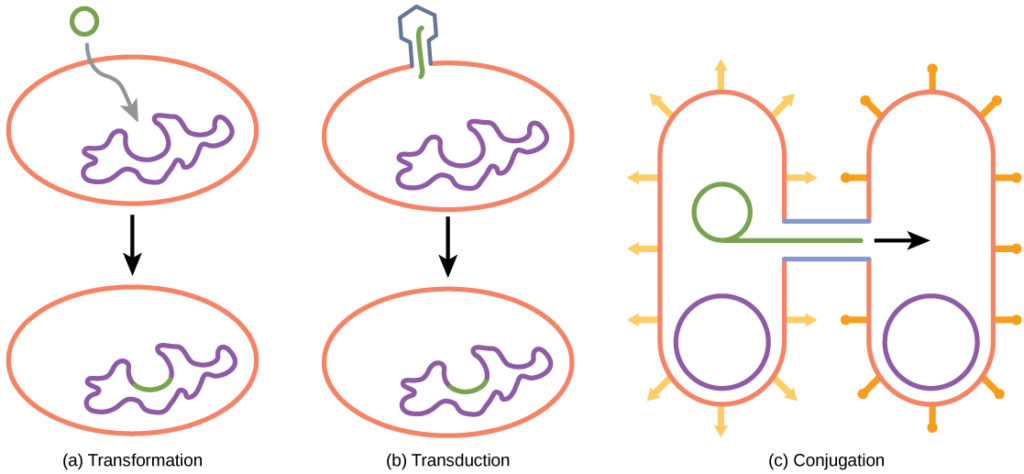
In transformation, the prokaryote takes in DNA found in its environment that is shed by other prokaryotes. If a nonpathogenic bacterium takes up DNA for a toxin gene from a pathogen and incorporates the new DNA into its own chromosome, it too may become pathogenic. In transduction, bacteriophages, the viruses that infect bacteria, sometimes also move short pieces of chromosomal DNA from one bacterium to another. Transduction results in a recombinant organism. Archaea are not affected by bacteriophages but instead have their own viruses that translocate genetic material from one individual to another. In conjugation, DNA is transferred from one prokaryote to another by means of a pilus, which brings the organisms into contact with one another. The DNA transferred can be in the form of a plasmid, a small circular piece of extrachromosomal DNA, or as a hybrid, containing both plasmid and chromosomal DNA. These three processes of DNA exchange are shown in Figure 2.
Reproduction can be very rapid: a few minutes for some species. This short generation time coupled with mechanisms of genetic recombination and high rates of mutation result in the rapid evolution of prokaryotes, allowing them to respond to environmental changes (such as the introduction of an antibiotic) very quickly.
How do scientists answer questions about the evolution of prokaryotes? Unlike with animals, artifacts in the fossil record of prokaryotes offer very little information. Fossils of ancient prokaryotes look like tiny bubbles in rock. Some scientists turn to genetics and to the principle of the molecular clock, which holds that the more recently two species have diverged, the more similar their genes (and thus proteins) will be. Conversely, species that diverged long ago will have more genes that are dissimilar.
Scientists at the NASA Astrobiology Institute and at the European Molecular Biology Laboratory collaborated to analyze the molecular evolution of 32 specific proteins common to 72 species of prokaryotes.[1] The model they derived from their data indicates that three important groups of bacteria—Actinobacteria, Deinococcus, and Cyanobacteria (which the authors call Terrabacteria)—were the first to colonize land. (Recall that Deinococcus is a genus of prokaryote—a bacterium—that is highly resistant to ionizing radiation.) Cyanobacteria are photosynthesizers, while Actinobacteria are a group of very common bacteria that include species important in decomposition of organic wastes.
The timelines of divergence suggest that bacteria (members of the domain Bacteria) diverged from common ancestral species between 2.5 and 3.2 billion years ago, whereas archaea diverged earlier: between 3.1 and 4.1 billion years ago. Eukarya later diverged off the Archaean line. The work further suggests that stromatolites that formed prior to the advent of cyanobacteria (about 2.6 billion years ago) photosynthesized in an anoxic environment and that because of the modifications of the Terrabacteria for land (resistance to drying and the possession of compounds that protect the organism from excess light), photosynthesis using oxygen may be closely linked to adaptations to survive on land.
Prokaryotes (domains Archaea and Bacteria) are single-celled organisms lacking a nucleus. They have a single piece of circular DNA in the nucleoid area of the cell. Most prokaryotes have a cell wall that lies outside the boundary of the plasma membrane. Some prokaryotes may have additional structures such as a capsule, flagella, and pili.
- Battistuzzi, FU, Feijao, A, and Hedges, SB. A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land. BioMed Central: Evolutionary Biology 4 (2004): 44, doi:10.1186/1471-2148-4-44. ↵
Contributors and Attributions
- Biology. Provided by: OpenStax CNX. Located at: http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8. License: CC BY: Attribution. License Terms: Download for free at http://cnx.org/contents/185cbf87-c72...f21b5eabd@10.8

