Study Questions
Answer questions 6.1 -6.3 using the following biochemical pathway for fruit color.Assume all mutations (lower case allele symbols) are recessive, and that either precursor 1 or precursor 2 can be used to produce precursor 3.If the alleles for a particular gene are not listed in a genotype, you can assume that they are wild-type.
6.1 If1 and 2 and 3 are all colorless, and 4 is red, what will be the phenotypes associated with the following genotypes?
- aa
- bb
- dd
- aabb
- aadd
- bbdd
- aabbdd
- What will be the phenotypic ratios among the offspring of a cross AaBb × AaBb?
- What will be the phenotypic ratios among the offspring of a cross BbDd× BbDd?
- What will be the phenotypic ratios among the offspring of a cross AaDd × AaDd?
6.2 If1 and 2 are both colorless, and 3 is blue and 4 is red, what will be the phenotypes associated with the following genotypes?
- aa
- bb
- dd
- aabb
- aadd
- bbdd
- aabbdd
- What will be the phenotypic ratios among the offspring of a cross AaBb × AaBb?
- What will be the phenotypic ratios among the offspring of a cross BbDd× BbDd?
- What will be the phenotypic ratios among the offspring of a cross AaDd × AaDd?
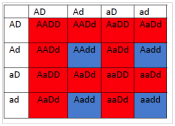
6.3 If1 is colorless, 2 is yellow and 3 is blue and 4 is red, what will be the phenotypes associated with the following genotypes?
- aa
- bb
- dd
- aabb
- aadd
- bbdd
- aabbdd
- What will be the phenotypic ratios among the offspring of a cross AaBb × AaBb?
- What will be the phenotypic ratios among the offspring of a cross BbDd× BbDd?
- What will be the phenotypic ratios among the offspring of a cross AaDd × AaDd?
6.4 Which of the situations in questions 6.1 – 6.3 demonstrate epistasis?
6.5 If the genotypes written within the Punnett Square are from the F2 generation, what would be the phenotypes and genotypes of the F1 and P generations for:
- Figure 6.6
- Figure 6.8
- Figure 6.10
- Figure 6.12
6.6 To better understand how genes control the development of three‐dimensional structures, you conducted a mutant screen in Arabidopsis and identified a recessive point mutation allele of a single gene (g) that causes leaves to develop as narrow tubes rather than the broad flat surfaces that develop in wild‐type (G). Allele g causes a complete loss of function. Now you want to identify more genes involved in the same process. Diagram a process you could use to identify other genes that interact with gene g. Show all of the possible genotypes that could arise in the Fgeneration.
6.7 With reference to question 6.6, if the recessive allele, g is mutated again to make allele g*, what are the possible phenotypes of a homozygous g* g* individual?
6.8 Again, in reference to question 6.7, what are the possible phenotypes of a homozygous aagg individual, where a is a recessive allele of a second gene? In each case, also specify the phenotypic ratios that would be observed among the F1 progeny of a cross of AaGg x AaGg
6.9 Calculate the phenotypic ratios from a dihybrid cross involving the two loci shown in Figure 6.13.There may be more than one possible set of ratios, depending on the assumptions you make about the phenotype of allele b.
6.10 Use the product rule to calculate the phenotypic ratios expected from a trihybrid cross.Assume independent assortment and no epistasis/gene interactions.
Chapter 6 - Answers
6.1 If 1 and 2 and 3 are all colorless, and 4 is red, what will be the phenotypes associated with the following genotypes? All of these mutations are recessive. As always, if the genotype for a particular gene is not listed, you can assume that alleles for that gene are wild-type.
- red(because A and B are redundant, so products 3 and then 4 can be made)
- red(because A and B are redundant, so products 3 and then 4 can be made)
- white (because product3 will accumulate and it is colorless)
- white (because only product 1 and 2 will be present and both are colorless)
- white (because only product 1 and 3 will be present and both are colorless)
- white (because only product 2 and 3 will be present and both are colorless)
- white (because only product 1 and 2 will be present and both are colorless)
- 15 red : 1 white
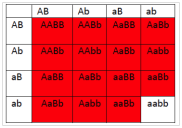
- 12 red : 4 white
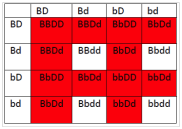
- 12 red :4 white
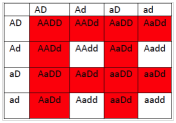
6.2
- red(because A and B are redundant, so products 3 and then 4 can be made)
- red(because A and B are redundant, so products 3 and then 4 can be made)
- blue (because product 3 will accumulate, and it is blue)
- white (because only product 1 and 2 will be present and both are colorless)
- blue (because only product 1 and 3 will be present and 1 is colorless and 3 is blue)
- blue(because only product 2 and 3 will be present and 2 is colorless and 3 is blue)
- white (because only product 1 and 2 will be present and both are colorless)
- 15 red : 1 white
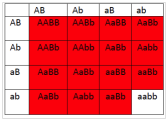
- 12 red : 4 blue
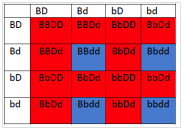
- 12 red : 4 blue

6.3
- red(because A and B are redundant, so products 3 and then 4 can be made)
- red(because A and B are redundant, so products 3 and then 4 can be made)
- blue (because product 3 will accumulate, and it is blue)
- yellow (because only product 1 and 2 will be present and 1 is colorless and 2 is yellow)
- blue (because only product 1 and 3 will be present and 1 is colorless and 3 is blue)
- green? (because only product 2 and 3 will be present and 2 is yellow and 3 is blue, so probably the fruit will be some combination of those two colors)
- yellow (because only product 1 and 2 will be present and 1 is colorless and 2 is yellow)
- 15 red : 1 yellow
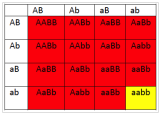
- 12 red : 3 blue:1 green

- 12 red : 4 blue
6.4 Epistasis is demonstrated when the phenotype for a homozygous mutant in one gene is the same as the phenotype for a homozygous mutant in two genes. So, the following situations from questions 6.1-6.3 demonstrated epistasis:
6.1 No epistasis is evident from the phenotypes, even though we know from the pathway provided that D is downstream of A and B.
6.2 The phenotypes show that D is epistatic to A, because aadd looks like AAdd or Aadd. Also D is epistatic to B, because bbdd looks like BBdd or Bbdd.
6.3 The phenotypes show that D is epistatic to A, because aadd looks like AAdd or Aadd. The phenotypes do not provide evidence for epistasis between B and D.
6.5 The answer is the same for a) – d)
P could have been either: AABB xaabboraaBB x AAbb;
F1 was : AaBb x AaBb
6.6 Conduct an enhancer/suppressor screen (which can also result in the identification of revertants, as well)
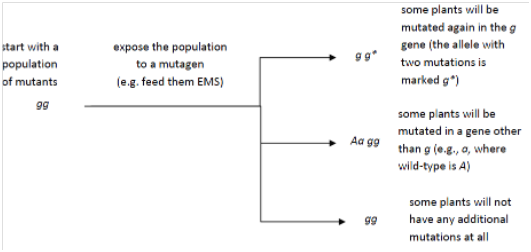
allow the plants to self‐pollinate in order to make any new, recessive mutations homozygous
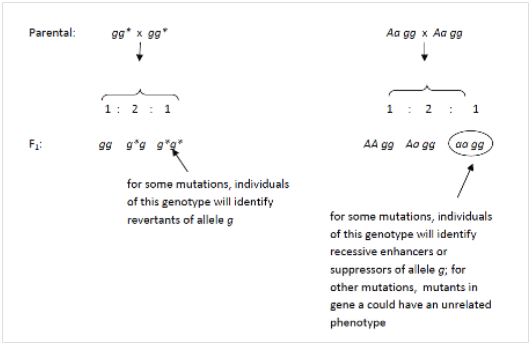
6.7 Depending which amino acids were altered, and how they were altered, a second mutation in g*g* could either have no effect (in which case the phenotype would be the same as gg), or it could possibly cause a reversion of the phenotype to wild‐type, so that g*g* and GG have the same phenotype.
6.8 Depending on the normal function of gene A, and which amino acids were altered in allele a, there are many potential phenotypes for aagg:
Case 1: If the normal function of gene A is in an unrelated process (e.g. A is required for
root development, but not the development of leaves), then the phenotype of aagg will
be: short roots and narrow leaves. The phenotypic ratios among the progeny of a
dihybrid cross will be:
| 9 | 3 | 3 | 1 |
| A_G_ | A_gg | aaG_ | aagg |
| wild-type | tubular leaves normal roots | short roots normal leaves | tubular leaves short roots |
Case 2: If the normal function of gene A is in the same process as G, such that a is a recessive allele that increases the severity of the gg mutant (i.e. a is an enhancer of g) then the phenotype of aagg could be : no leaves. The phenotypic ratios among the progeny of a dihybrid cross depend on whether aa mutants have a phenotype independent of gg, in other words, do aaG_ plants have a phenotype that is different from wild‐type or from A_gg. There is no way to know this without doing the experiment, since it depends on the biology of the particular gene, mutation and pathway involved, so there are three possible outcomes:
Case 2a) If aa is an enhancer of gg, and aaG_ plants have a mutant phenotype that differs from wild‐type or (A_gg) then the phenotypic ratios among the progeny of a dihybrid cross will be:
| 9 | 3 | 3 | 1 |
| A_G_ | A_gg | aaG_ | aagg |
| wild-type | tubular leaves (some phenotype that differs from gg; maybe small twisted leaves) | abnormal leaves | no leaves |
Case 2b) If aa is an enhancer of gg, and aaG_ plants have a mutant phenotype that is the same as A_gg , the phenotypic ratios among the progeny of a dihybrid cross will be:
| 9 | 6 | 1 |
| A_G_ | A_gg aaG_ | aagg |
| wild-type | tubular leaves | no leaves |
Case 2c) If aa is an enhancer of gg, and aaG_ do not have a phenotype that differs from wild‐type then the phenotypic ratios among the progeny of a dihybrid cross will be:
| 12 | 3 | 1 |
| A_G_ aaG_ | A_gg | aagg |
| wild-type | tubular leaves | no leaves |
Case 3: If the normal function of gene A is in the same process as G, such that a is a recessive allele that decreases the severity of the gg mutant (i.e. a is an suppressor of g) then the phenotype of aagg could be : wild‐type. The phenotypic ratios among the progeny of a dihybrid cross depend on whether aa mutants have a phenotype independent of gg, in other words, do aaG_ plants have a phenotype that is different from wild‐type or from A_gg. There is no way to know this without doing the experiment, since it depends on the biology of the particular gene, mutation and pathway involved, so there are three possible outcomes:
Case 3a) If aa is a suppressor of gg, and aaG_ plants have a mutant phenotype that differs from wild‐type or (A_gg) then the phenotypic ratios among the progeny of a dihybrid cross will be:
| 10 | 3 | 3 |
| A_G_ aagg | A_gg | aaG_ |
| wild-type | tubular leaves (some phenotype that differs from gg) | no leaves |
Case 3b) If aa is an suppressor of gg, and aaG_ plants have a mutant phenotype that is the same as A_gg the phenotypic ratios among the progeny of a dihybrid cross will be:
| 10 | 6 |
| A_G_ aagg | A_gg aaG_ |
| wild-type | tubular leaves |
Case 3c) If aa is an suppressor of gg, and aaG_ plants do not have a phenotype that differs from wild‐type then the phenotypic ratios among the progeny of a dihybrid cross will be:
| 13 | 3 |
| A_G_ aaG_ aagg | A_gg |
| wild-type | tubular leaves |
Case 4: If the normal function of gene A is in the same process as G, such that a is a
recessive allele that with a phenotype that is epistatic to the gg mutant then the
phenotype of both aaG_ and aagg could be : no leaves. The phenotypic ratios among
the progeny of a dihybrid cross will be:
| 9 | 4 | 3 |
| A_G_ | aaG_ aagg | A_gg |
| wild-type | no leaves | tubular leaves |
Case … ?: There are many more phenotypes and ratios that could be imagined (e.g.
different types of dominance relationships, different types of epistasis, lethality…etc).
Isn’t genetics wonderful? It is sometimes shocking that more people don’t want to
become geneticists.
The point of this exercise is to show that many different ratios can be generated,
depending on the biology of the genes involved. On an exam, you could be asked to
calculate the ratio, given particular biological parameters. So, this exercise is also meant
to demonstrate that it is better to learn how to calculate ratios than just trying to
memorize which ratios match which parameters. In a real genetic screen, you would
observe the ratios, and then try to deduce something about the biology from those
ratios.
6.9 Calculate the phenotypic ratios from a dihybrid cross involving the two loci shown in Figure 6.13. There may be more than one possible set of ratios, depending on the assumptions you make about the phenotype of allele b.
Assuming that bb has no phenotype on its own (i.e. A_bb looks like A_B_), then aaB_ will have the mutant phenotype, and A_bb, A_B_, and aabb will appear phenotypically wild-type. The phenotypic ratio will be 13 wild-type : 3 mutant.
6.10 For a dihybrid cross, there are 4 classes, 9:3:3:1. In a trihybrid cross without gene interactions, each of these 4 classes will be further split into a 3:1 ratio based on the phenotype at the third locus. For example, 9 x 3 =27 and 9 x 1 = 9. This explains the first two terms of the complete ratio: 27:9:9:9:3:3:3:1.














