2.63: Sample Project Report
- Page ID
- 102001
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)*** DO NOT COPY / COPY & EDIT THIS SAMPLE FOR YOUR REPORT***
All text in this report will be flagged by TurnItIn as not your words (i.e. plagarism). Even if you change some words here and there, TurnItIn knows that these are not your words.
Identification of two Bacterial Species in a Mixed Culture Using Traditional Microbiological Identification Approaches
Written By: Rosanna Hartline
Introduction
Bacterial Diversity & Importance of Species Identification
Bacteria are among the most diverse types of life on Earth (Locey and Lennon, 2016). There are billions, perhaps trillions, of bacterial species occupying habitats that are living, such as animals, and non-living habitats with a vast variety of temperatures, pH, and salinities (Parker et al., 2022). The metabolic and physical and adaptations enabling bacterial survival is wide-ranging, particularly given that these microscopic organisms are only a single cell big (Parker et al., 2022). The impacts bacteria have on other living things and the environment are extensive. Bacteria are responsible for critical nutrient cycling activities in environmental systems that would not occur without them (Parker et al., 2022). Without bacteria, nutrient availability on Earth would be highly limited and greatly reduce the amount of life that could exist on this planet (Parker et al., 2022).
Bacteria live symbiotically with other living things in parasitic, commensal, and mutualistic ways (Parker et al., 2022). In fact, living in and on a single human, there are more than 100 trillion bacterial cells (Locey and Lennon, 2016). To understand the interactions of each bacterial species and their impacts on the environment and other living things, bacterial species must be identified and characterized. The implications of the types of bacterial species living in different habitats and environments ranges from destructive and lethal effects of some bacterial species to bacterial roles and activities that are crucial for the existence of life. In this study, the goal was to apply traditional microbiological approaches to successfully identify bacterial species of unknown identifies in a mixed culture. To accomplish this, the following approaches were used: quadrant streak plate, Gram stain, starch hydrolysis test, H2S production test, oxidase test, and lactose fermentation test.
Quadrant Streak Plate
To identify bacterial species using traditional microbiological methods, isolating a single bacterial species from other microbes is crucial. Subsequent microbiological tests will have indeterminant results if there are multiple microbial species mixed, since there is no way of knowing which species is producing any positive test result. To isolate bacterial species, a commonly used approach is the quadrant streak plate technique (Raymond et al., 2022). This approach utilizes a petri plate with nutrient-rich media and four streak zones (Raymond et al., 2022). The first zone will have the densest growth since it is streaked directly from the source of the microbes being examined. The following zones are pulled from the streak immediately before, and the microbes present on the inoculating loop are killed by flaming before each subsequent streak. The result is dilution of the number of microbes in each streak, until the fourth quadrant produces locations on the petri plate where only single cells are deposited on the petri plate agar (Hartline, 2022). A bacterial colony will develop from a single cell as the cells divide by binary fission, and thereby produce an isolated colony that will contain only one bacterial species (Klamm, 2019). These isolated colonies can then be characterized, aiding in species identification, and then may be transferred to a separate culture to create a monoculture for subsequent identification tests. Qualities such as colony color, form, size, and appearance are useful information to distinguish between different species of bacteria. These can be determined by careful examination of isolated colonies on a petri plate.
Gram Stain
The diversity of bacterial species in the world is subdivided into two major groups based on their cell wall structures and are designated as either Gram-positive or Gram-negative based on the results of the Gram stain (Raymond et al., 2022). The Gram stain will stain Gram-positive bacteria purple since the purple-colored crystal violet stain is not removed from the Gram-positive cell wall during the decolorization step of the Gram stain due to the thick peptidoglycan layer in the cell wall outside of the plasma membrane (Parker et al., 2022). The Gram-positive bacterial cell wall also contains teichoic acid and does not have an outer membrane (Hartline, 2022). For Gram-negative bacteria, the purple-colored crystal violet stain is readily removed from its thin peptidoglycan layer during the decolorization step of the Gram stain and will appear pink due to the counterstain safranin used after the decolorization step (Raymond et al., 2022). Most species that stain Gram-negative have a thin peptidoglycan layer outside the of plasma membrane with an outer membrane outside of the peptidoglycan layer that has embedded lipopolysaccharide (also known as endotoxin) (Hartline, 2022; Parker et al., 2022).
Starch Hydrolysis Test
One of the many microbiological tests used to characterize bacterial species and aid in species identification is the starch hydrolysis test. Starch is a polysaccharide, and therefore is composed of many sugars that bacteria can use as an energy source and a carbon source (Parker et al., 2022). If a bacterial species has the amylase gene, it can produce the enzyme amylase enabling it to hydrolyze (break down) starch and utilize this rich energy and carbon source (Hartline, 2022). To determine if bacteria hydrolyze starch, bacteria are grown on a starch plate and following incubation, iodine is added to the plate. Iodine reacts with starch to produce a blue-black color enabling visualization of starch and where it is and is not present in the petri plate agar (Pakpour and Horgan 2021). If starch has been broken down by the bacteria, the agar will lack the blue-black color around the bacterial growth indicating that starch has been hydrolyzed, since starch is absent near the bacteria (Pakpour and Horgan 2021).
H2S Production
Use of SIM deep medium can be used to determine multiple characteristics of a bacterial species in a single test tube, including determining if bacteria can produce H2S (Pakpour and Horgan 2021). Production of H2S can indicate that either a bacterial species can convert cysteine (an amino acid) to pyruvate or that a bacterial species conducts a form of anaerobic respiration where H2S is produced when a sulfur-containing compound is used as a final electron acceptor (Hartline, 2022). The SIM medium contains ferrous sulfide and if bacteria produce H2S, the ferrous sulfide will react with H2S to produce a black precipitate, indicating the bacterial species is H2S-positive (Pakpour and Horgan 2021). The absence of black precipitate characterizes the bacterial species as H2S-negative, since it does not produce H2S gas.
Oxidase Test
Cytochrome c oxidase is an enzyme that some bacteria use as part of their electron transport chain in aerobic respiration (Raymond et al., 2022). Only some bacterial species have the cytochrome c oxidase gene and therefore produce this enzyme, making it a useful characteristic for differentiating bacterial species in the identification process (Raymond et al., 2022). Some bacterial species do not have the cytochrome c oxidase enzyme, but they can still conduct aerobic respiration using different enzymes (Hartline, 2022). Testing for the presence or absence of this enzyme is not an indicator of whether a species conducts aerobic respiration or not, just whether the species uses the cytochrome c oxidase as the enzyme that transfers electrons from the electron transport chain to oxygen, the final electron acceptor in this process (Hartline, 2022). The oxidase test determines whether a bacterial species has the cytochrome c oxidase enzyme. This test uses a chemical called tetramethyl-p-phenylenediamine and if cytochrome c oxidase is present in the bacteria, it will remove electrons from this chemical and the product molecule is a blueish or purplish color (Klamm, 2019). Therefore, oxidase positive bacteria produce a blueish or purplish color in the first 30 seconds of mixing the chemical with live bacteria and oxidase negative bacteria do not produce this color change.
Lactose Fermentation Test
Some bacterial species can use fermentation as a method of producing ATP and some bacterial species are not. Further, the types of sugars that different bacterial species can ferment also differs based on the genes (and therefore enzymes) that the species have or do not have (Hartline, 2022). The lactose fermentation test determines whether bacterial species are capable of utilizing lactose, a type of sugar, to conduct fermentation and will indicate whether a bacterial species will make acid, gas, or both as byproducts of the fermentation process (Lee 2021). Bacterial species that metabolize lactose have the gene for the enzyme beta-galactosidase enabling the breakdown lactose, a disaccharide, into monosaccharides that can enter catabolic pathways such as fermentation (Hartline 2022). To conduct this test, a lactose-containing medium with a pH indicator and Durham tube is inoculated with the bacteria being tested. If acid is produced by the fermentation process, the medium will change from red to yellow (Lee, 2021). If gas is produced by the fermentation process, a pocket of gas will form inside of the Durham tube. If no lactose fermentation occurs, no acid or gas will be produced.
Experimental Overview
Bacterial identification is an important skill for people in a vast range of professions where bacteria can impact their work including healthcare, food safety, and water safety. This experiment aimed to separate the bacterial species in a mixed culture and utilized traditional microbiological identification approaches to identify the bacterial species present in that culture. These techniques resulted in successful identification of Bacillus subtilis as one of the species in that mixed culture. The other species in the culture was Escherichia coli but was misidentified in this process as Serratia marcescens.
Methods
Quadrant Streak Plate
From a TSB culture containing a mixture of two unknown bacterial species, a loopful of culture was aseptically transferred to a single streak along one side of a petri plate. The loop was flamed and cooled before an edge of the initial streak was dragged into a second streak on the petri plate (Hartline, 2022). The loop was again flamed, cooled, and part of the second streak was dragged into the third streak (Hartline, 2022). This was repeated one more time to complete a fourth streak (Hartline, 2022). This process was repeated eight times to produce a total of eight quadrant streak plates. The plates were inverted and incubated at 30 °C for 48 hours.
Gram Stain
Bacterial smears were prepared by either by aseptically transferring several loops of liquid culture to a microscope slide or aseptically transferring a bacterial colony using a loop from a petri plate to a drop of saline on a microscope slide. The slide was placed onto a slide warmer until dry and then heat-fixed by passing the slide through a Bunsen burner flame three times. Crystal violet was applied to stain the smear for one minute, rinsed with deionized water, covered with Gram’s iodine for one minute, rinsed with deionized water, decolorized with ethanol for five to fifteen seconds, rinsed with deionized water, counterstained with safranin for one minute, and rinsed with deionized water (Hartline, 2022). Following this staining procedure, the slide was blotted with bibulous paper and examined with a light microscope.
Starch Hydrolysis Test
A loop was used to aseptically transfer isolated bacteria from a TSA slant culture to create a single-line streak on the surface of a petri plate with starch agar medium. The plate was inverted and incubated at 37 °C for 48 hours. The plate was then flooded with iodine to visualize presence and absence of starch in the agar.
H2S Production
An inoculation needle was used to aseptically transfer bacteria from a TSA slant to a SIM deep culture and incubated at 37 °C for 48 hours.
Oxidase Test
A sterile plastic loop was used to aseptically transfer bacteria from the SIM deep culture to a moistened oxidase test strip. Results of the test were interpreted within the first 30 seconds. This test was repeated a total of three times to ensure that results were accurate.
Lactose Fermentation Test
Bacteria were aseptically transferred from a TSA slant to a test tube with phenol red lactose medium containing a Durham tube. This culture was incubated at 37 °C for 12 days.
Results & Discussion
Initial Gram Stain of Mixed Bacterial Culture
A Gram stain of the mixed culture of unknown bacteria designated as ‘H’ revealed two distinctly different bacterial species since one stained purple and one stained pink. Both species exhibited rod-shaped cells and as a result are designated as bacilli.
Pink-colored cells indicated these cells were a Gram-negative species and therefore likely have a cell wall containing a thin layer of peptidoglycan and an outer membrane with embedded lipopolysaccharide (Parker et al., 2022). The crystal violet primary stain was removed from the Gram-negative cells during the decolorization step of the Gram stain due to the thin layer of peptidoglycan being unable to retain the stain. These cells appeared pink due to the counterstain safranin.
The purple-colored cells suggested these were a Gram-positive species that have a thick layer of peptidoglycan in its cell walls resulting in the crystal violet stain being retained during the decolorization step of the Gram stain (Parker et al., 2022).
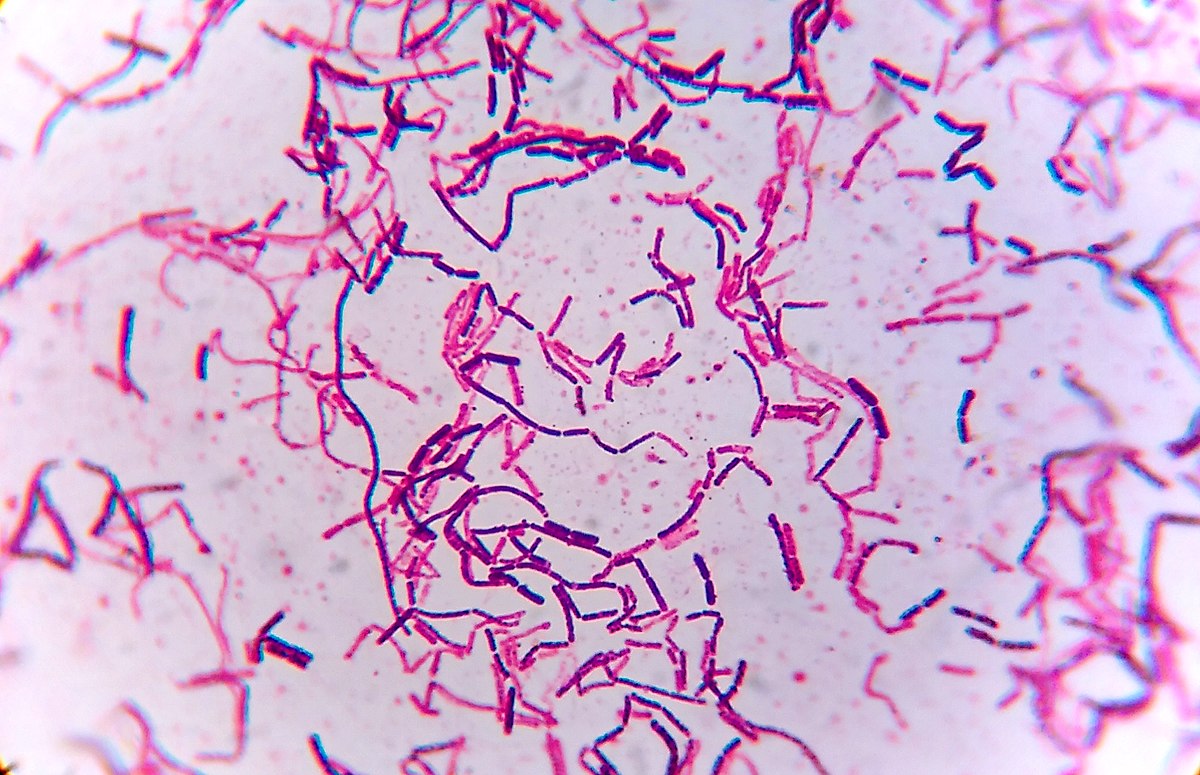
Figure 1: Gram stain of mixed bacterial culture ‘H’ imaged at 1000x magnification.
The initial Gram stain of the mixed bacterial culture ‘H’ revealed that the two unknown bacterial species were a Gram-positive bacillus species and a Gram-negative bacillus species.
Isolation of Bacterial Species
As a result of the quadrant streak plate procedure, isolated colonies were successfully produced. There were two distinctly different types of colonies that were differentiated by shape, color, and size. These differences in colony characteristics suggest that these are different microbial species (Figure 2).

Figure 2: The quadrant streak plate from the mixed culture of unknown bacterial designed ‘H.’
One of the colony types, hereafter designed as bacterial specie ‘alpha,’ were a bright ivory color and shiny (Hartline, 2022). The colonies measured between 1 mm and 3 mm in diameter and had a circular form with sharp edges and raised elevation.
The other colony type, designated as ‘beta,’ produced dry and dull looking colonies with a milky-yellow coloration, an irregular form, flat elevation, and measured between 3 mm and 5 mm in diameter (Hartline, 2022). These colonies had distinct color variations that occurred as irregular, wavy rings within the colonies.
Identification of Bacterial Species Alpha
Gram Stain
A Gram stain conducted on the unknown isolated bacterial species Alpha revealed that this species has rod-shaped cells, or bacilli, and the cells appeared pink after the Gram stain (Figure 3).
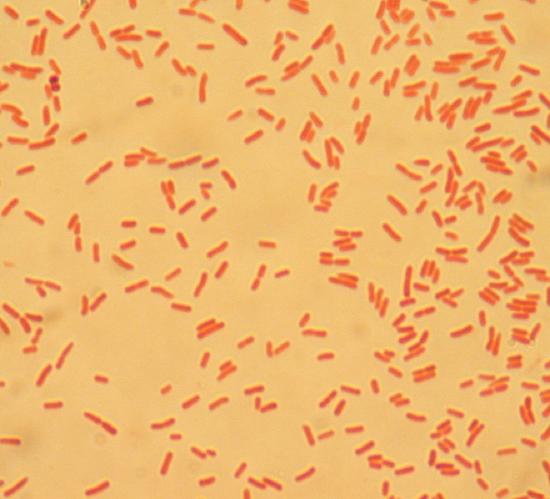
Figure 3: Isolated unknown bacterial species Alpha after Gram stain at 1000x magnification.
Since unknown bacterial species Alpha produced pink-colored cells in the Gram stain, this indicates it is a Gram-negative species (Hartline, 2022). This species likely has a cell wall with a thin layer of peptidoglycan outside of the plasma membrane and an outer membrane containing lipopolysaccharide (Hartline, 2022; Parker et al., 2022).
H2S Test
Since unknown bacterial species Alpha was Gram-negative, the next indicated test to conduct was to assess whether this species produces H2S gas or not. After inoculation and incubation of a SIM deep, the medium did not produce a black coloration (Figure 4).

Figure 4: SIM deep culture with unknown bacterial species Alpha.
Since the SIM deep did not produce a black coloration, this means that no H2S was produced in the culture to interact with the ferrous sulfide in the medium (Pakpour and Horgan 2021). Therefore, unknown bacterial species Alpha does not produce H2S and is H2S-negative. This result reveals that unknown bacterial species Alpha does not produce pyruvate from cysteine and does not conduct the form of anaerobic respiration that produces H2S gas as a byproduct (Hartline, 2022).
Although not directly a part of this identification protocol, the presence or absence of motility can be determined using this SIM deep culture (Pakpour and Horgan 2021). Growth migrated away from the stab line in the SIM deep suggesting unknown bacterial species Alpha is motility positive and therefore has one or more flagella enabling movement in this bacterial species (Hartline, 2022; Pakpour and Horgan 2021).
Oxidase Test
Due to the H2S negative test result, the identification protocol next indicates the oxidase test. The oxidase test did not produce a purple or blue color in the first 30 seconds (Figure 5). The test was repeated a total of three times to ensure the validity of this result and the results did not change.

Figure 5: One of the test strips produced from the oxidase test of unknown bacterial species Alpha.
The lack of purple or blue color in under 30 seconds means that unknown bacterial species Alpha is oxidase negative. These results indicate that unknown bacterial species Alpha does not have the cytochrome c oxidase enzyme or the gene that codes for cytochrome c oxidase (Raymond et al., 2022). While this result does not indicate whether this bacterial species can conduct aerobic respiration or not, it does indicate that it does not use the cytochrome c oxidase enzyme to transfer electrons from the electron transport chain to O2 (Hartline, 2022). If this species does conduct aerobic respiration, it uses a different enzyme for this process at the end of the electron transport chain (Hartline, 2022).
Lactose Fermentation Test
Since the oxidase test produced a negative result for unknown bacterial species Alpha, the next indicated identification test in the protocol was the lactose fermentation test. After inoculation and incubation, the phenol red lactose medium maintained a red color and the Durham tube was still filled with the liquid medium and lacked a gas pocket inside of it (Figure 6).

Figure 6: Lactose fermentation culture of unknown bacterial species Alpha.
Since the phenol red lactose medium did not become yellow, this indicates that acid was not produced by the bacteria. No air pocket in the Durham tube in this culture indicates that no gas was produced by the bacteria in this culture. Since neither gas nor acid were produced, this indicates that bacterial species Alpha is lactose fermentation negative. This result suggests this bacterial species either does not conduct fermentation, does not have the beta-galactosidase gene for breaking down lactose, or both (Hartline, 2022).
Based on this result, unknown bacterial species Alpha was identified using this laboratory protocol as Serratia marcescens.
Identification of Bacterial Species Beta
Gram Stain
The isolated bacterial species designated as Beta was Gram-stained and results show the cells appear as purple rods (Figure 7).

Figure 7: Gram stain of unknown bacterial species Beta magnified at 1000x.
The purple coloration of this bacterial species after the Gram stain indicates this species is Gram-positive. This result indicates this species has a thick layer of peptidoglycan in its cell wall that retains the purple coloration of the crystal violet stain during the Gram stain protocol (Parker et al., 2022). The rod-shaped cells indicate this speices have cells that are Gram-positive bacilli.
Starch Hydrolysis Test
Since unknown bacterial species Beta stained Gram-positive, the next indicated test in the identification protocol was the starch hydrolysis test. Following incubation of the unknown bacteria on a starch plate and staining the starch plate with iodine, a clear zone surrounded the bacterial growth on the petri plate (Figure 8).
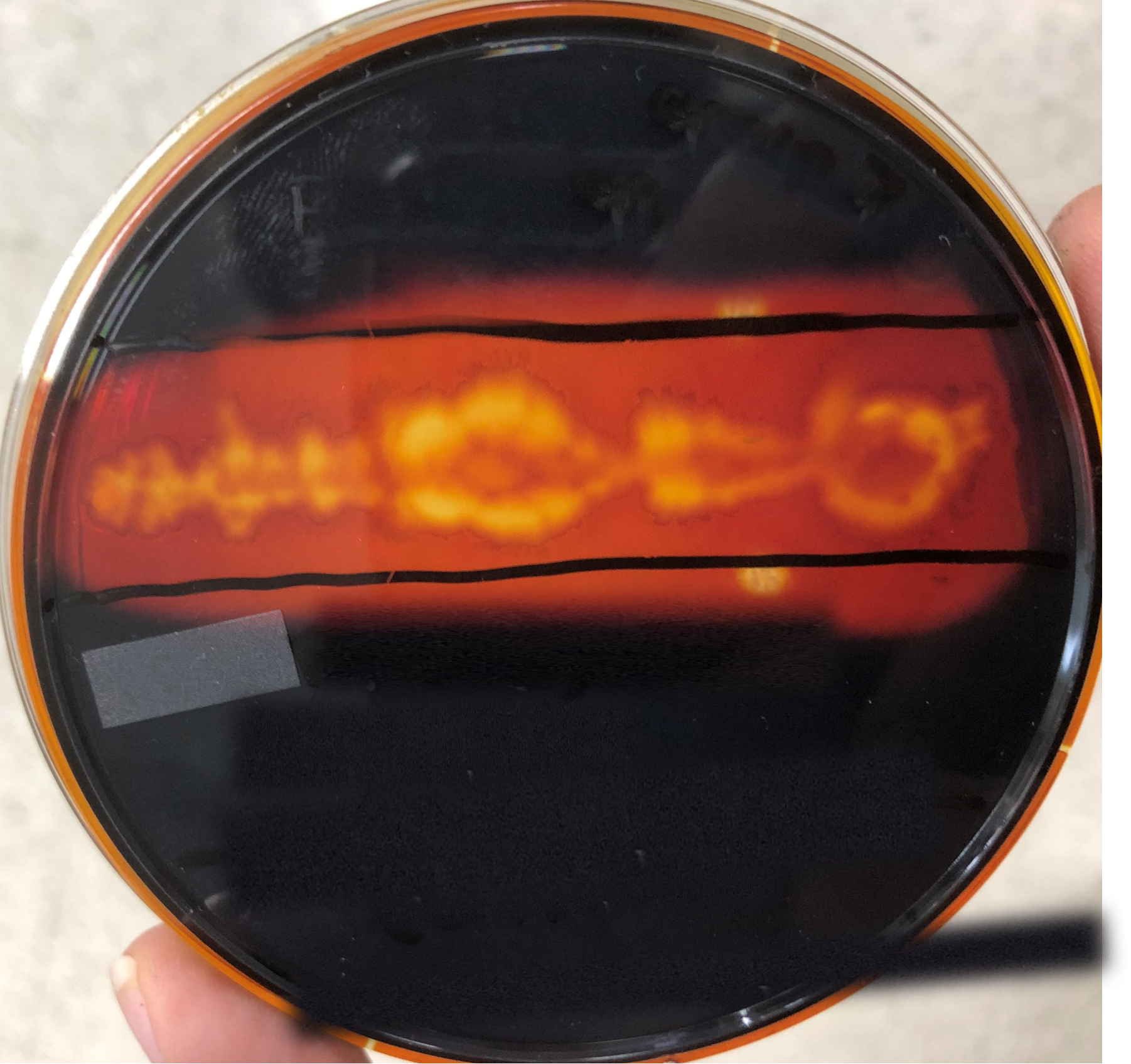
Figure 8: Starch hydrolysis plate flooded with iodine with growth of unknown bacterial species Beta.
The starch hydrolysis test showed a clear zone surrounding the bacterial growth indicating that starch was absent in the agar surrounding unknown bacterial species Beta (Pakpour and Horgan 2021). This result reveals that Beta is starch hydrolysis positive and therefore must have the amylase gene to produce the amylase enzyme that breaks down starch (Hartline, 2022).
Since the starch hydrolysis test was positive for unknown bacterial species Beta, the identification protocol indicates that this species is Bacillus subtilis.
Conclusion
Unknown bacterial species Alpha was identified as Serratia marcescens using this identification protocol using traditional microbiological techniques. In fact, this species was not S. marcescens, but was Escherichia coli indicating one or more errors occurred resulting in incorrect identification of this bacterial species. There are two plausible explanations for this misidentification. First, it is possible when unknown bacterial species Alpha was aseptically collected from the TSA slant that the loop was too hot, and any bacteria collected from the slant were killed by the heat of the loop. As a result, the lactose fermentation tube may not have been successfully inoculated, resulting in the lactose fermentation negative result. The other possible explanation for incorrect identification is that a contaminant colony on the quadrat streak plate was collected and tested throughout this identification process instead of the actual unknown bacterial species that was targeted.
Unknown bacterial species Beta was identified as Bacillus subtilis in this experiment. The Gram-positive bacterial species in unknown culture ‘H’ was indeed B. subtilis and therefore identification of this species was successful. This result was likely due to several factors including careful aseptic technique to prevent contamination, thoughtful examination and documentation of each laboratory result throughout the process, and careful execution of all the laboratory protocols to ensure accurate experimental results.
Works Cited
Hartline R. 2022. Microbiology Laboratory Manual. LibreText. Retrieved from: https://bio.libretexts.org/Courses/West_Hills_College_-_Lemoore/Microbiology_Laboratory_Manual
Klamm LS. 2019. Klamm’s Microbiology Laboratory Manual. MOspace Institutional Repository. Retrieved from: https://mospace.umsystem.edu/xmlui/handle/10355/69341
Lee, 2021. MB352 General Microbiology Laboratory 2021 (Lee). LibreText. Retrieved from: https://bio.libretexts.org/Courses/North_Carolina_State_University/MB352_General_Microbiology_Laboratory_2021_(Lee)
Locey KJ, Lennon JT. 2016. Scaling laws predict global microbial diversity. Proceedings of the National Academy of Sciences of the United States of America. 113: 5970-5975.
Pakpour N, Horgan S. 2021. General Microbiology Lab Manual (Pakpour & Horgan). LibreText. Retrieved from: https://bio.libretexts.org/Learning_Objects/Laboratory_Experiments/Microbiology_Labs/Book%3A_General_Microbiology_Lab_Manual_(Pakpour_and_Horgan)
Parker N, Schneegurt M, Thi Tu A-H, Lister P, Forster BM, Allen S, Auman A, Brelles-Mariño G, Alhadeff Feldman M, Flowers P, Pinchuk G, Rowley B, Sutherland M, Franklund C, Paterson A. 2022. Microbiology. OpenStax. Retrieved from: https://openstax.org/details/books/microbiology
Raymond J, Boorse G, Mason A. 2022. Red Mountain Microbiology. Maricopa Community Colleges. Retrieve from: https://open.maricopa.edu/redmountainmicro/
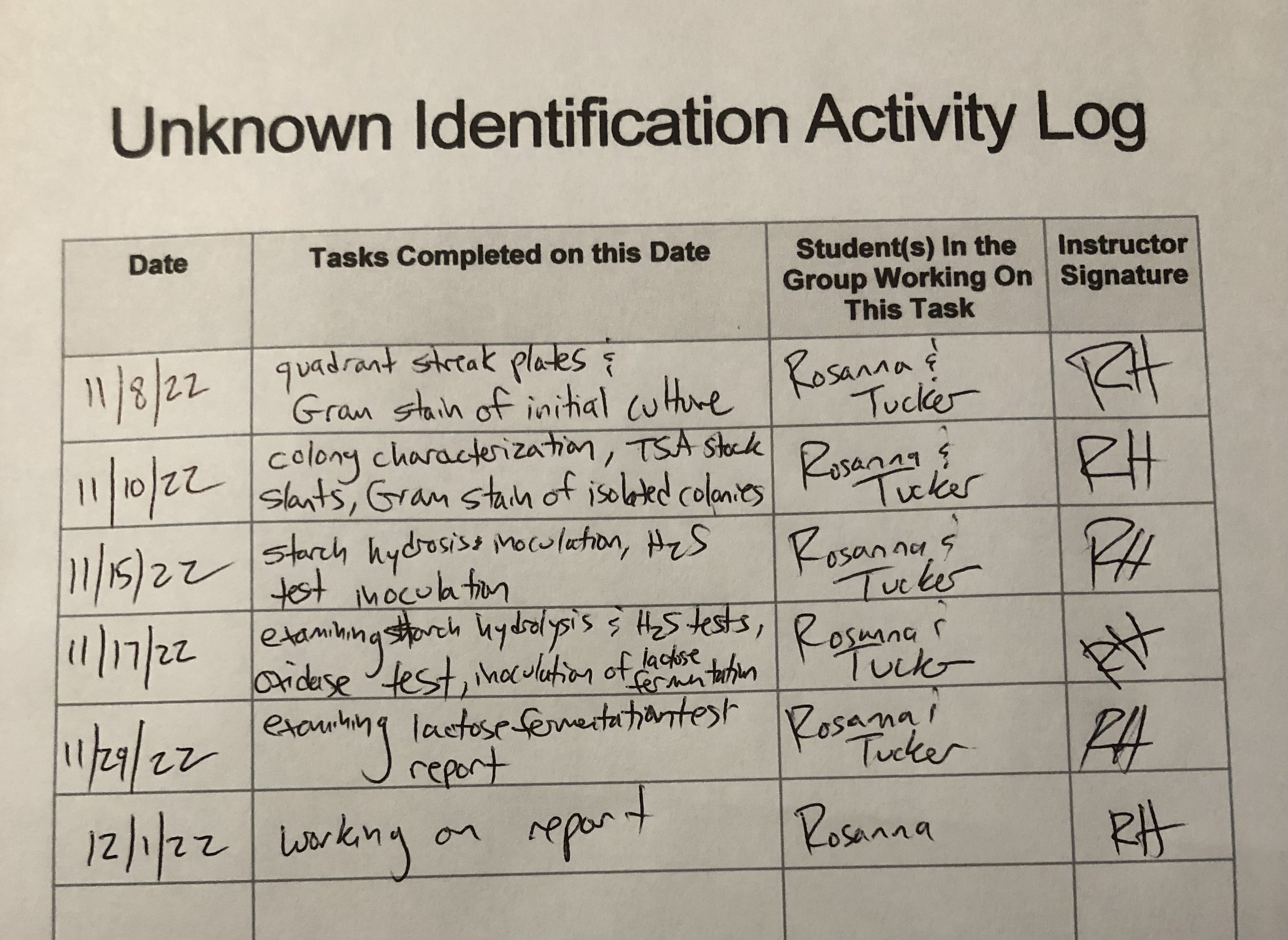
Activity Log
Attributions*
- E choli Gram.JPG by Bobjgalindo is licensed under CC BY-SA 4.0
- Gram positive bacilli.jpg by Microrao is licensed under CC BY-SA 4.0
- Red Mountain Microbiology by Jill Raymond Ph.D.; Graham Boorse, Ph.D.; Anne Mason M.S. is licensed under CC BY-NC 4.0
- Yersinia enterocolitica in SIM Agar1 125.jpg by A doubt is licensed under CC BY-SA 4.0
*Attributions are not a part of the sample Unknown Identification Project Report and are not to be included in student reports.


