18: Genetic Engineering
- Page ID
- 10730
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Genetic engineering is the deliberate manipulation of DNA, using techniques in the laboratory to alter genes in organisms. Even if the organisms being altered are not microbes, the substances and techniques used are often taken from microbes and adapted for use in more complex organisms.
Steps in Cloning a Gene
Let us walk through the basic steps for cloning a gene, a process by which a gene of interest can be replicated many times over. Let us pretend that we are going to genetically engineer E. coli cells to glow in the dark, a characteristic that they do not naturally possess.
- Isolate DNA of interest – first we need to identify the genes or genes that we are interested in, the target DNA. If we want our E. coli cells to glow in the dark, we need to find an organism that possesses this trait and identify the gene or genes responsible for the trait. The green fluorescent protein (GFP) commonly used as an expression marker in molecular techniques was originally isolated from jellyfish.In cloning a gene it is helpful to use a cloning vector, typically a plasmid or virus, capable of independent replication that will stably carry the target DNA from one location to another. Plasmid vectors are available from both bacteria and yeast.
- Cut DNA with restriction endonucleases – once the target and vector DNA have been identified, both types of DNA are cut using restriction endonucleases. These enzymes recognize short sequences of DNA that are 4-8 bp long. The enzymes are widespread in both bacteria and archaea, with each enzyme recognizing a specific inverted repeat sequence that is palindromic (reads the same on each DNA strand, in the 5’ to 3’ direction).
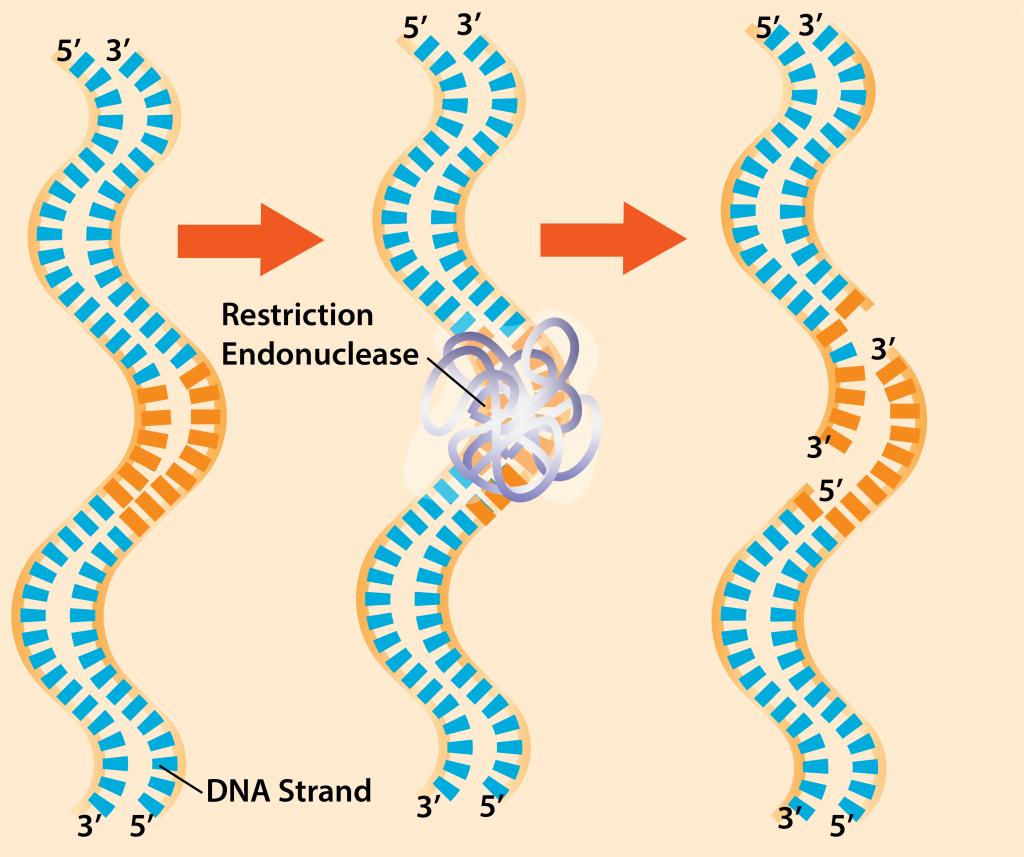
Restriction Endonucleases.
While some restriction endonucleases cut straight across the DNA (i.e. blunt cut), many make staggered cuts, producing a very short region of single-stranded DNA on each strand. These single-stranded regions are referred to as “sticky ends,” and are invaluable in molecular cloning since the unpaired bases will recombine with any DNA having the complementary base sequence.
- Combine target and vector DNA – after both types of DNA have been cleaved by the same restriction endonuclease, the two types of DNA are combined together with the addition of DNA ligase, an enzyme that repairs the covalent bonds on the sugar-phosphate backbone of the DNA. This results in the creation of recombinant DNA, DNA molecules that contain the DNA from two or more sources, also known as chimeras.
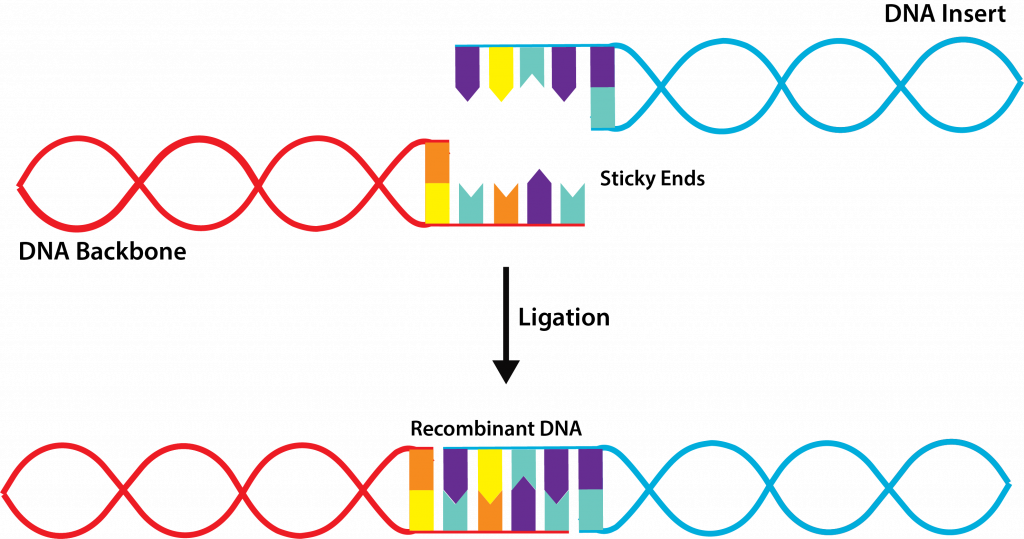
Ligation.
- Introduce recombined molecule into host cell – once the target DNA has been stably combined with vector DNA, the recombinant DNA must be introduced into a host cell, in order for the genes to be replicated or expressed. There are different methods for introducing the recombinant DNA, largely depending upon the complexity of the host organism. In the case of bacteria, transformation is often the easiest method, using competent cells to pick up the recombinant DNA molecules. Alternatively, electroporation can be used, where the cells are exposed to a brief pulse of high –voltage electricity causing the plasma membrane to become temporarily permeable to DNA passage.While some cells will acquire recombinant DNA with the appropriate configuration (i.e. target DNA combined with vector DNA), the method also will yield cells carrying recombinant DNA with alternate DNA combinations (i.e. plasmid DNA combining with another plasmid DNA molecule or target DNA attached to more target DNA). The mixture is referred to as a genomic library and must be screened to select the appropriate clone. If random fragments of DNA were originally used (instead of isolation of the appropriate target DNA genes), the process is referred to as shotgun cloning and can yield thousands or tens of thousands of clones to be screened.
Introducing recombinant DNA into cells other than bacteria
Agrobacterium tumefaciens and the Ti plasmid

Tumor-Inducing Plasmid.
Agrobacterium tumefaciens is a plant pathogen that causes tumor formation called crown gall disease. The bacterium contains a plasmid known as the Ti (tumor inducing) plasmid, which inserts bacterial DNA into the host plant genome. Scientists utilize this natural process to do genetic engineering of plants by inserting foreign DNA into the Ti plasmid and removing the genes necessary for disease, allowing for the production of transgenic plants.
Gene gun
A gene gun uses very small metal particles (microprojectiles) coated with the recombinant DNA, which are blasted at plant or animal tissue at a high velocity. If the DNA is transformed or taken up by the cell’s DNA, the genes are expressed.
Viral vectors
For a viral vector, virulence genes from a virus can be removed and foreign DNA inserted, allowing the virus capsid to be used as a mechanism for shuttling genetic material into a plant or animal cell. Marker genes are typically added that allow for identification of the cells that took up the genes.
DNA techniques
Gel Electrophoresis
Gel electrophoresis is a technique commonly used to separate nucleic acid fragments based on size. It can be used to identify particular fragments or to verify that a technique was successful.
A porous gel is prepared made of agarose, with the concentration adjusted based on expected size. Nucleic acid samples are deposited into wells in the gel and an electrical current is applied. Nucleic acid, with its negative charge, will move towards the positive electrode, which should be placed at the bottom of the gel. The nucleic acid will move through the gel, with the smallest pieces encountering the least resistance and thus moving through the fastest. The length of passage of each nucleic acid fragment can be compared to a DNA ladder, with fragments of known size.
Polymerase Chain Reaction (PCR)
The polymerase chain reaction or PCR is a method used to copy or amplify DNA in vitro. The process can yield a billionfold copies of a single gene within a short period of time. The template DNA is mixed with all the ingredients necessary to make DNA copies: primers (small oligonucleotides that flank the gene or genes of interest by recognizing sequences on either side of it), nucleotides (the building blocks of DNA), and DNA polymerase. The steps involve heating the template DNA in order to denature or separate the strands, dropping the temperature to allow the primers to anneal, and then heating the mixture up to allow the DNA polymerase to extend the primers, using the original DNA as an initial template. The cycle is repeated 20-30 times, exponentially increasing the amount of target DNA in a few hours.

Polymerase Chain Reaction (PCR). By Enzoklop (Own work) [CC BY-SA 3.0], via Wikimedia Commons
Uses of Genetically Engineered Organisms
There can be numerous reasons to create a genetically modified organism (GMO) or transgenic organism, defined as a genetically modified organism that contains a gene from a different organism. Typically the hope is that the GMO will provide needed information or a product of value to society.
Source of DNA
Genetically engineered organisms can be made so that a piece of DNA can be easily replicated, providing a large source of that DNA. For example, a gene associated with breast cancer can be spliced into the genome of E. coli, allowing for the rapid production of the gene so that it may be sequenced, studied, and manipulated, without requiring repeated tissue donations from human volunteers.
Source of RNA
Antisense RNA is ssRNA that is complementary to the mRNA that will code for a protein. In cells it is made as a way to control target genes. There has been increasing interest in the use of antisense RNA as a way to prevent diseases that are caused by the production of a particular protein.

Antisense RNA. By Robinson R [CC BY 2.5], via Wikimedia Commons
Source of Protein
Since microbes replicate so rapidly, it can be extremely advantageous to use them to manufacture proteins of interest or value. Given the right promoters, bacteria will express genes for proteins that are not naturally found in bacteria, such as cytokine. Genetically engineered cells have been used to make a wide variety of proteins of use to humans, such as insulin or human growth hormone.

Key Words
genetic engineering, cloning, target DNA, green fluorescent protein (GFP), cloning vector, restriction endonuclease, sticky ends, DNA ligase, recombinant DNA, chimera, transformation, electroporation, genomic library, shotgun cloning, Agrobacterium tumefaciens, Ti plasmid, gene gun, microprojectiles, viral vector, gel electrophoresis, DNA ladder, polymerase chain reaction (PCR), template DNA, primer, nucleotide, DNA polymerase, denaturing, annealing, extending, genetically modified organism (GMO), transgenic organisms, antisense RNA.
Study Questions
- Define genetic engineering.
- Identify and describe the basic steps used in the genetic engineering of a bacterial cell. What components are needed and why?
- Summarize the different ways that recombinant DNA can be inserted into a cell or organism. Be able to provide specific examples.
- Describe techniques used in the manipulation of DNA. What are the essential components of each process?
- Explain the different applications of genetic engineering.


