17: Microbial Genetics
- Page ID
- 10723
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Let’s talk about sex. Bacterial sex. Ha! That is going to be difficult, since bacteria do not have sex. Which presents a real problem for bacteria (and archaea, too) – how do they get the genetic variability that they need? They might need a new gene to break down an unusual nutrient source or degrade an antibiotic threatening to destroy them – acquiring the gene could mean the difference between life and death. But where would these genes come from? How would the bacteria get a hold of them? We are going to explore the processes that bacteria use to acquire new genes, the mechanisms known as Horizontal Gene Transfer (HGT).
Conjugation
Conjugation is the process by which a donor bacterium transfers a copy of a plasmid to a recipient bacterium, through a pilus. The process requires cell-to-cell contact. The donor cell (F+) has a conjugative plasmid, an extrachromosomal piece of dsDNA that codes for the proteins necessary to make a threadlike filament known as a pilus. The pilus is used to bind to the recipient (F-) cell, bringing it in close proximity to the donor cell. It is believed that a channel is then opened between the two cells, allowing for a ssDNA copy of the plasmid to enter the recipient cells. Both cells then make the complementary copy to the ssDNA, resulting in two F+ cells capable of conjugation.
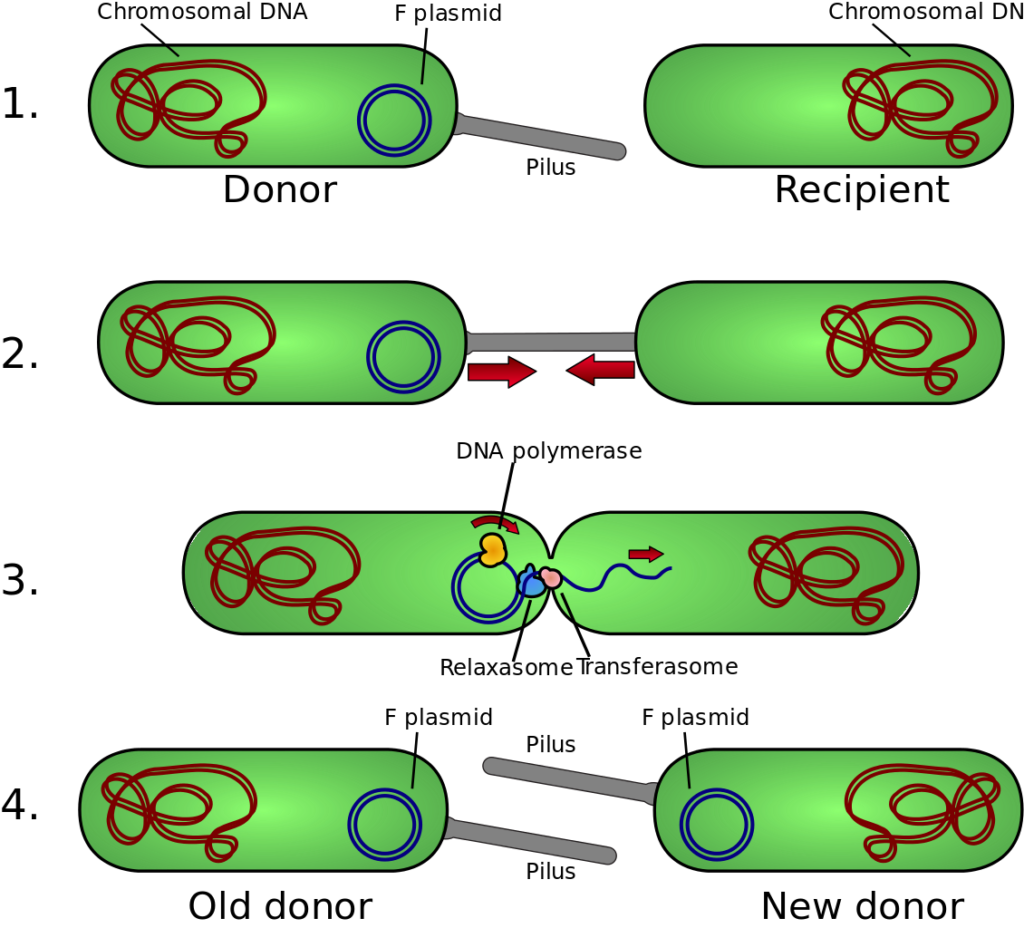
Conjugation. By Adenosine (Own work) [CC BY-SA 3.0], via Wikimedia Commons
Transformation
The process of transformation also allows a bacterial cell to acquire new genes, but it does not require cell-to-cell contact. In this process the new genes are acquired directly from the environment. Typically the process requires a donor cell that at some point lysed and released naked DNA to the environment. The recipient cell is one that is capable of taking up the DNA from the environment and incorporating it into its own genome, where the cell is described as being competent.
There are mechanical and chemical means of encouraging a cell to pick up DNA from the environment, but natural competence is determined genetically. The process typically occurs at the end of exponential phase of growth or beginning of the stationary phase, in the presence of high cell density and limited nutrients. Under these conditions specific proteins are manufactured including DNA-binding proteins (DNA translocase), endonucleases, and transmembrane channel proteins. Gram negative cells also make a cell wall autolysin, to transport the DNA across the outer membrane.
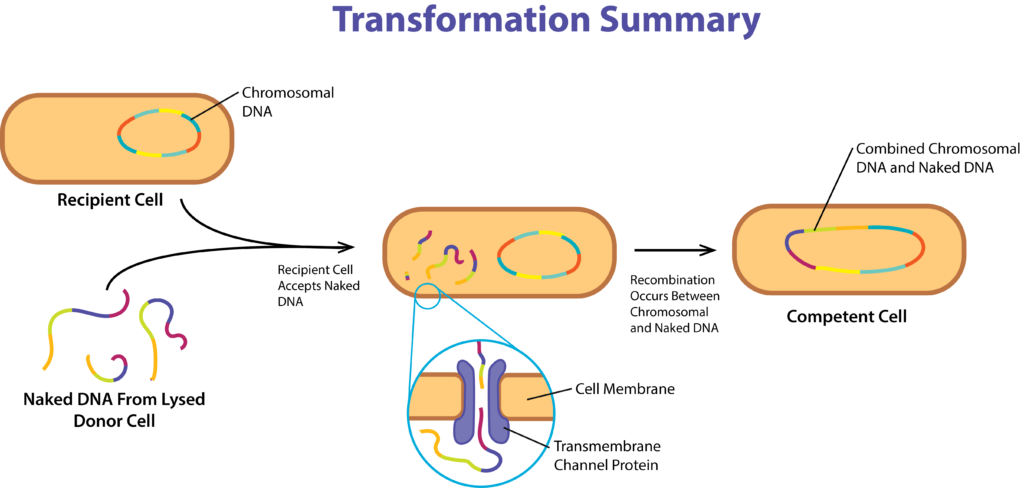
Gene Acquisition via Transformation.
Random pieces of DNA bind to receptors on the outside of the cell and are then transported into the cell by the DNA translocase, through the transmembrane channel, a large structure often involving numerous different proteins. An endonuclease can be used to degrade one strand of dsDNA, if only ssDNA may pass into the cell, or to cleave the DNA fragment into smaller sizes .Once inside the cell, the DNA must be incorporated into the bacterial chromosome by RecA (see Molecular Recombination below), for the genes to be expressed.
Transduction
Transduction involves the use of a virus, a bacteriophage, to act as a conduit for shuttling bacteria genes from one cell to another, thus negating the necessity for cell-to-cell contact. There are two different types of transduction: generalized transduction and specialized transduction.
Generalized Transduction
In generalized transduction, a bacterial host cell is infected with either a virulent or a temperate bacteriophage engaging in the lytic cycle of replication. After the first three steps of replication (absorption, penetration, and synthesis), the virus enters into the assembly stage, during which fully formed virions are made. During this stage, random pieces of bacterial DNA are mistakenly packaged into a phage head, resulting in the production of a transducing particle. While these particles are not capable of infecting a cell in the conventional sense, they can bind to a new bacterial host cell and inject their DNA inside. If the DNA (from the first bacterial host cell) is incorporated into the recipient’s chromosome, the genes can be expressed.
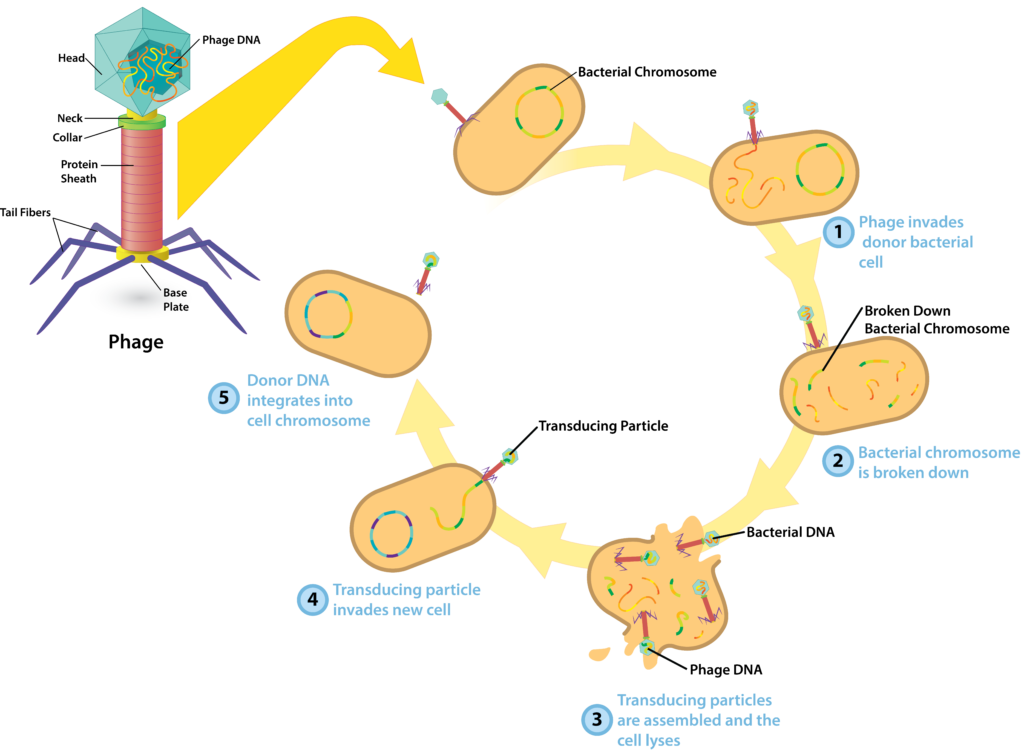
Generalized Transduction.
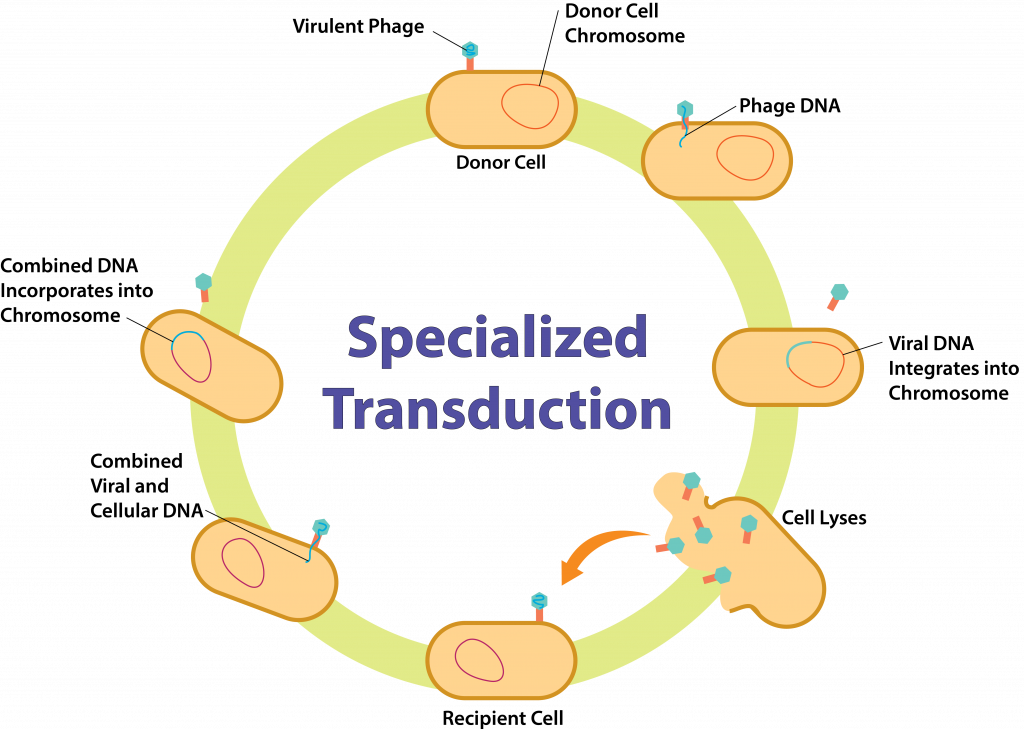
Specialized Transduction
Specialized transduction can only occur with temperate bacteriophage, since it involves the lysogenic cycle of replication. The bacteriophage randomly attaches to a bacterial host cell, injecting viral DNA inside. The DNA integrates into the chromosome of the host cell, forming a prophage. At some point induction occurs, where the prophage is excised from the bacterial chrosomsome. In specialized transduction, the excision is incorrectly performed and a portion of bacterial genes immediately adjacent to the viral genes are excised too. Since this DNA is used as the template for the synthesis stage, all copies will be a hybrid of viral and bacterial DNA, and all resulting virions will contain both viral and bacterial DNA.
Once the cell is lysed, the virions are released to infect other bacterial host cells. Each virion will attach to the host cell and inject in the DNA hybrid, which could be incorporated into the host chromosome, if a prophage is formed. At this point the second bacterial host cell can contain its own DNA, DNA from the previous bacterial host cell, and viral DNA.
Molecular Recombination
In each of the cases of HGT, the process is only successful if the genes can be expressed by the altered cell. In conjugation, the genes are located on a plasmid, under the control of promoters on the plasmid. In transformation and transduction, where naked DNA is gaining access to the cell, the DNA could easily be broken down by the cell with no genetic expression occurring. In order for the genes to be expressed, the DNA must be recombined with the recipient’s chromosome.
The most common mechanism of molecular recombination is homologous recombination, involving the RecA protein. In this process DNA from two sources are paired, based on similar nucleotide sequence in one area. An endonuclease nicks one strand, allowing RecA to pair up bases from different strands, a process known as strand invasion. The cross-over between DNA molecules is resolved with resolvase, which cuts and rejoins the DNA into two separate dsDNA molecules.
Recombination can also occur using site-specific recombination, a process often used by viruses to insert their genome into the chromosome of their host. This type of recombination is also used by transposable elements (see next section).
Transposable Elements
Finally, we shouldn’t leave the topic of microbial genetics without at least exploring the role of transposable elements or “jumping genes.” While these can play a very big role in the activation and inactivation of bacterial genes, the best explanation derives from the work of Barbara McClintock in corn, who won the Nobel Prize for her research in 1983. She demonstrated that transposable elements can be responsible for the activation or inactivation of genes within an organism.
Transposable elements are relatively simple in structure, designed to move from one location to another within a DNA molecule by a process known as transposition. All transposable elements code for the enzyme transposase, the enzyme responsible allowing transposition to occur, and have short inverted repeats (IRs) at each end.
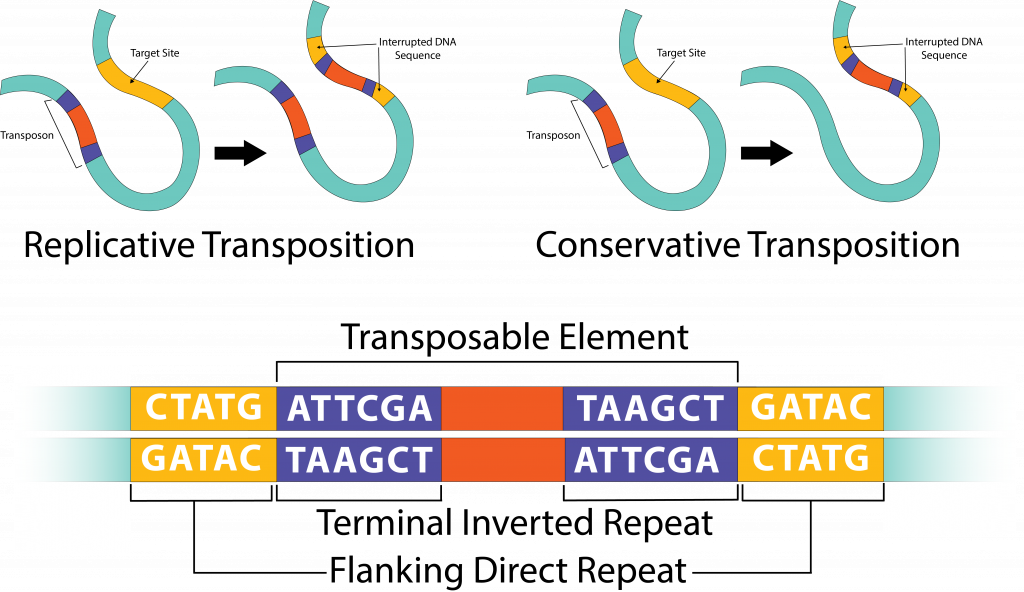
Transposition.
The simplest transposable element is an insertion sequence (IS), which contains the transposase and IRs of varying lengths. A transposontypically contains additional genes, with the exact type varying widely from transposon to transposon. A transposon can be removed from one location and relocated to another (the cut-and-paste model), a process known as conservative transposition. Alternatively, it can be copied, with the copy being inserted at a second site, in a process known as replicative transposition.
Key Words
Horizontal Gene Transfer (HGT), conjugation, donor, recipient, conjugative plasmid, F-, F, transformation, naked DNA, competence, competent cell, DNA translocase, endonuclease, autolysin, RecA, transduction, generalized transduction, transducing particle, specialized transduction, molecular recombination, homologous recombination, resolvase, site-specific recombination, transposable elements, transposition, transposase, inverted repeats (IR), insertion sequence (IS), transposon, conservative transposition, replicative transposition.
Essential Questions/Objectives
- What is horizontal gene transfer? What are the three mechanisms for this to occur in bacteria?
- What are the components needed for the processes of transformation, conjugation, and transduction? How does each process occur? What genes are involved in each process?
- How do generalized and specialized transduction differ? What is the end result of each?
- What is recombination? What is the importance to bacteria & archaea?
- What are the two types of recombination? What are the details of each type? What components are needed for each type?
- What is transposition? What is required for the process to occur? What is a transposable element?
Exploratory Questions (OPTIONAL)
- How could transposons be used in the study of bacterial genetics?


