Case Study: Unusual microbes
- Page ID
- 404
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Each of the organisms discussed here challenges our simple views of the microbial world. Many are accommodated easily enough by simply broadening our view a bit. Some may perhaps raise some important questions. Most of these organisms were discovered recently (or at least the key points of interest were discovered recently). That should be a clue that more surprises are likely.
- Introduction
- Big bacteria
- Small bacteria
- Bacteria that give birth to live young
- Square bacteria
- Microbes with too much DNA
- Microbes with too many genes
- Bacteria with "atypical" chromosomes
- Bacteria that can count -- and talk
- How hot?
- Bacteria that know where north is
- Bacteria that eat other bacteria
- Multicellular bacteria
- A huge virus
- The biggest microbe?
- Briefly noted
- More "Curious microbes"
Introduction
This page was written in collaboration with Borislav Dopudja, a third-year science student at the University of Zagreb. It grew out of some casual but extensive discussions we were having exploring some of the oddities of the microbial world, things that don't quite fit with our common views. It seems worthwhile to share some of these. There is no attempt here to be profound, but rather to have some fun enjoying the diversity of the microbial world. Borislav's web site is: www.pluff-sky.net/. I have also listed it, with more information, on my page of Internet resources: Biology Miscellaneous in the Biology: other section.
Terminology
What are "microbes"? Microbes (or microorganisms) are small organisms. For our purposes, that means single-celled organisms. The single cells may be prokaryotic or eukaryotic. The prokaryotic microbes include the bacteria and the archaea (or the eubacteria and archaebacteria, by older terminology). The eukaryotic microbes include the protists (protozoa), the fungi and at least the unicellular algae. The terms are not always used consistently, especially in older literature. Whether viruses should be included is a matter of taste, and I won't be entirely consistent there; for the most part, we will discuss cellular organisms.
Sources
Most of the links below are to web sites that are suitable for "the general audience". A few links to articles from the regular scientific literature are given in small type. In particular, in some cases I have included links to the first reports of these organisms or of key features.
Many links are to Microbe magazine -- or to its precursor, ASM News. Microbe is the news magazine of the American Society for Microbiology. Microbe is written for microbiologists, but written to be enjoyed by a wide range of non-specialists. Both the news stories and feature articles can serve well as readable material with serious scientific content, yet not too technical. Microbe is now freely available online. Links to individual items are given as they come up. If you would like to browse Microbe magazine -- recommended! -- go to http://www.microbemagazine.org/. The current issue will come up; for more, see "Explore Microbe" at the left.
Some links are given to original articles or news stories in Science magazine. Some of these are freely available online, though you may need to create a free registration before getting access to the full text. In general, Science releases research articles -- but not news stories -- for free access 12 months after publication; their file goes back to about 1997. (Those with institutional subscription access, such as those using university computers at UC Berkeley, have full access, and will not be asked to register or log on.) The home page for Science magazine: http://www.sciencemag.org.
Big bacteria
One of the most characteristic properties of bacteria is that they are small. Microscopic. Barely visible under the microscope: we can tell their general shape, but can generally see very little structure. Typical dimensions are on the order of 1 micrometer (1 μm). So, have a look at Epulopiscium and Thiomargarita -- bacteria big enough to be seen with the naked eye. These bacteria -- at least the larger specimens -- approach 1 millimeter (1 mm) in size. One of these was first reported in 1985 -- but not understood to be a bacterium until 1993; the other was first reported in 1999.
Epulopiscium
Epulopiscium grows in the gut of certain fish. It has a complex life cycle, which is coordinated with the daily rhythm of its host. This complex -- and unusual -- life cycle qualifies Epulopiscium for another section of this page: Bacteria that give birth to live young.
- http://microbewiki.kenyon.edu/index.php/Epulopiscium. From the Microbe Wiki.
- www.microbelibrary.org/index....ium-fishelsoni. From the Microbe Library at ASM.
- www.accessexcellence.org/LC/ST/st12bg.php. "Epulopiscium fishelsoni, Big bug baffles biologists!", an essay on this unusual bug, from Peggy E Pollak & W Linn Montgomery, both early investigators of Epulos. (Another Access Excellence page is listed on this page, in the section The biggest microbe?. The Access Excellence site is listed as a general resource on my page of Miscellaneous Internet Resources, under Of local interest... -- since it had its origins near here.)
Cornell researchers study bacterium big enough to see -- the Shaquille O'Neal of bacteria. Press release (May 6, 2008) on new work showing that an Epulopiscium cell contains 100,000 or so copies of its genome, thus has many times more DNA than a human cell. This site is worth it for the pictures alone. The upper picture is a classic, showing an Epulo, a paramecium and an ordinary E. coli bacterium. http://www.news.cornell.edu/stories/...cteria.kr.html. The paper, from Angert's lab at Cornell and collaborators in Australia and New Zealand, is: J E Mendell et al, Extreme polyploidy in a large bacterium. PNAS 105:6730-6734, 5/6/08. Online: http://www.pnas.org/content/105/18/6730.abstract.
The first report of Epulopiscium, describing it as a large and peculiar cigar-shaped organism, presumably a protist: L Fishelson et al, A unique symbiosis in the gut of tropical herbivorous surgeonfish (Acanthuridae: Teleostei) from the Red Sea. Science 229:49, 7/5/85. The abstract is freely available at http://www.sciencemag.org/content/22...08/49.abstract. You may or may not be able to get the full article at that site. If not and you have an institutional subscription to JStor, such as at UCB, try www.jstor.org/stable/1695432.
The definitive report that Epulopiscium is really a bacterium: E R Angert et al, The largest bacterium. Nature 362:239, 3/18/93. http://www.nature.com/nature/journal.../362239a0.html.
Thiomargarita
Thiomargarita can be quite big, but it "cheats". It is mostly vacuole. Why? Well, it is quite like a deep sea diver carrying an oxygen tank. Thiomargarita uses nitrate ions in its respiration, rather than oxygen gas; the vacuole is a supply of nitrate that lets the bug continue to respire at great depths.
Is Life Thriving Deep Beneath the Seafloor? An article from the Woods Hole Oceanographic Institute (WHOI). http://www.whoi.edu/oceanus/viewArticle.do?id=2497. To focus on Thiomargarita, scroll down to "The world's largest bacterium". The article is by WHOI oceanographer Carl Wirsen, April 2004. WHOI microbiologist Andreas Teske was part of the team that discovered Thiomargarita; he is a co-author of the Science paper listed below as the original report.
| H N Schulz, Thiomargarita namibiensis: Giant microbe holding its breath. ASM News 68:122, 3/02. The news magazine ASM News -- now called Microbe -- is free online; this item is at newsarchive.asm.org/mar02/feature2.asp (HTML) or newsarchive.asm.org/mar02/images/f2.pdf (PDF). The figure at the right is "Figure 1" from that article. Note the scale bar (which is for part "A" of the Figure); the cells shown are over a half millimeter across. | 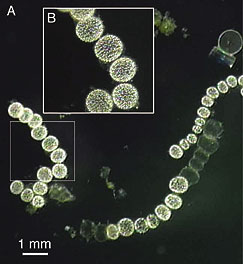 |
The original report on Thiomargarita: H N Schulz et al, Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284:493, 4/16/99. It is accompanied by a news story: B Wuethrich, Microbiology: Giant sulfur-eating microbe found. Science 284:415, 4/16/99. The article is freely available at: http://www.sciencemag.org/content/28...3/493.abstract.
Small bacteria
Scientists from UC Berkeley, led by Dr Jill Banfield, have found an archaeon smaller than any cellular organism previously known. It is about 200 nm (0.2 μm) diameter. It is so small that it is very near the "limit" of what people think might be the smallest possible organism. In fact, some people think it might be below that limit! Time will tell whether the new claim is valid. An important issue is whether this is a "complete" organism, or a parasite of some kind that is absolutely dependent on other cells to provide basic functions. This is part of their work on the acidic mine drainage from the Richmond Mine at Iron Mountain, Calif.
Shotgun sequencing finds nanoorganisms. A news release from UC Berkeley, December 2006, on this discovery: http://www.berkeley.edu/news/media/releases/2006/12/21_microbes.shtml. The original report on this tiny organism: B J Baker et al, Lineages of acidophilic archaea revealed by community genomic analysis. Science 314:1933, 12/22/06. http://www.sciencemag.org/content/31.../1933.abstract.
There is another story of small bacteria, a story that has been around for several years but has not really been confirmed. The basic idea is a claim that there are tiny bacteria involved in such processes as calcification of your arteries. These bacteria, which have been termed nanobacteria, are alleged to be even smaller than those discussed above -- far below any reasonable limit of what is "possible" for a living cell. Since these alleged organisms really do not fit in any modern understanding of what cells are, solid evidence is needed -- and is lacking. Two new papers appeared in early 2008 with rather strong evidence that these "things" are not alive. They appear to be some calcium minerals, complexed with protein. They may well be interesting, and they may still be involved in disease processes, but they are not bacteria. The Wikipedia entry is a good introduction to these "nanobacteria" (or "calcifying nanoparticles"), including the uncertainties that surround them. It notes these 2008 papers, and has links to them, and to one good news story on the new findings. http://en.Wikipedia.org/wiki/Nanobacterium.
Bacteria that give birth to live young
Bacteria divide by binary fission: they grow bigger, and then divide in two. But there are exceptions. An interesting type of exception occurs when bacteria seem to give birth to live young. That is, they develop new cells inside, and then liberate these daughter cells. One of the first cases where this type of bacterial reproduction was seen was with Epulopiscium, discussed in the section on Big bacteria. Then it was found in the bacterium Metabacterium -- but in a form that was easier to understand. It has long been known that some bacteria make spores. Specifically, bacteria of the genera Bacillus and Clostridia make "endospores": each cell makes one spore, a resistant structure that is capable of long term survival. Such spore formation does not increase the population, because each cell makes one spore. It merely results in a new type of cell, the resistant spore. But Metabacterium makes multiple spores per cell -- and rarely undergoes the more "ordinary" process of binary fission. Thus a variation of ordinary endospore formation has become the primary means of reproduction. With Epulopiscium, it would seem that this process has been modified further, so that what is produced is not spores but rather ordinary cells -- baby cells.
Thus both Metabacterium and Epulopiscium "give birth to live young" -- a process that can be thought of as a variation of ordinary endospore formation.
Esther Angert, Beyond binary fission: Some bacteria reproduce by alternative means. Microbe 1:127, 3/06: forms.asm.org/microbe/index.asp?bid=41230 (HTML) or forms.asm.org/ASM/files/ccLib...0306000127.pdf (PDF). Angert, at Cornell, works with both Metabacterium and Epulopiscium. She was the first to recognize that Epulopiscium was actually a bacterium.
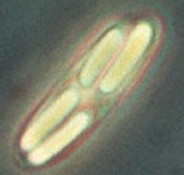
Metabacterium with four daughter spores. The figure at the right shows a single Metabacterium polyspora cell containing four spores, with the bright appearance that is typical of bacterial endospores. The individual spores are several μm long. The figure is a trimmed version of a figure at: author.cals.cornell.edu/cals/...abacterium.cfm. That page discusses the life cycle of Metabacterium, and how it relates to the natural environment for this organism. It is part of Esther Angert's web site; the home page is author.cals.cornell.edu/cals/...-lab/intro.cfm.
More links for Epulopiscium are in the section on Big bacteria.
Square bacteria
What is the shape of bacteria? Round. Or roundish -- such as rods with rounded ends. Certainly not square, with sharp corners. Imagine then the surprise of the scientist who, in 1980, found square bacteria with sharp corners, in concentrated salt solutions. They are not only square, but very thin -- about 200 nm (0.2 μm) thick. They seem to grow as two dimensional objects, increasing the size of their squares, but not their thickness. Square bacteria caught in the act of division look like a sheet of postage stamps.
Their thinness increases their surface to volume ratio; this may be important in helping them to maintain a proper intracellular environment. They probably spend much energy pumping ions out!
It is not known why they are square or how they achieve their squareness. The square bacteria are archaea. (That was recognized in the original report; at that time, the archaebacteria were considered a type of bacteria, whereas we now consider them a distinct group from the bacteria per se.) They have been named Haloquadratum walsbyi.

The following two links both include good pictures of the square bacteria. The figure at the right is a variation of one shown at the second site, Dyall-Smith's web site.
- Square bacteria grown in the laboratory. Press release, from the University of Melbourne, announcing the first successful growth of the square bacteria in the lab, October 2004. uninews.unimelb.edu.au/news/1855/.
- Web site for Dr Mike Dyall-Smith, group leader for that work: http://www.haloarchaea.com/. The broad topic of the site is Haloarchaea and Haloviruses.
Edwin Abbott would have loved them. http://www.ibiblio.org/eldritch/eaa/FL.HTM. A good read!
Genome paper: H Bolhuis et al, The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics 7:169, 7/4/06. Free online: http://www.biomedcentral.com/1471-2164/7/169.
The original report of these organisms: A E Walsby, A square bacterium. Nature 283:69, 1/3/80. It's a delightful little paper, a brief report of a quite unexpected observation. Online: http://www.nature.com/nature/journal.../283069a0.html. Even reading the opening lines, which are freely available there, gives a nice hint of the literary quality of this paper. However, it probably requires subscription for full access.
Microbes with too much DNA
The organism with the largest known genome? Amoeba dubia, a protist. 670 billion base pairs of DNA. That is about 200 times more than we have. What is the significance of this finding? It's not at all clear. In fact, it is hard to even find the source of the number. Is this really a measure of the haploid genome size? Or is it simply based on the cellular DNA content, with the assumption that the cell is diploid? Not only is the original source hard to pin down, there seems to be no modern work following it up. But the number is oft-quoted, so we quote it too. Someday we may understand what it means.
For a nice discussion of genome sizes, see http://www.genomesize.com/statistics.php. Scroll down to the section "A comment on the overall animal range", and what follows, including a nice graph summarizing genome sizes over all types of organisms. This is from T Ryan Gregory, Univ Guelph. Regardless of the ultimate verdict on the Amoeba dubia genome, many organisms have genomes much larger than ours.
In the web page referred to above, Gregory gives genome size in picograms (pg). Biologists often give genome sizes in base pairs (bp). 1 picogram of DNA is about 109 (one billion) base pairs. For example, the human genome contains about 3.5 billion bp, and weighs about 3.5 pg.
Gregory's genome size site is also referred to on my Internet resources - Molecular Biology page, under Genomes, and in the Musings post Who is #1: the most DNA? (March 7, 2011).
Microbes with too many genes
The human genome project brought us the revelation that we have only about 22,000 genes -- not all that many more than a worm (Caenorhabditis elegans, 20,000 genes) or a fruit fly (Drosophila melanogaster, 14,000 genes). Now, Trichomonas vaginalis - a common sexually-transmitted protist (protozoan)... Its genome sequence was reported in January 2007. Preliminary analysis suggests about 60,000 genes.
Don't make too much of this. Gene counts are notoriously difficult, as we learned from the human genome project. Identification of genes simply by looking at DNA sequences is something of an art. In fact, the report offers multiple numbers for the gene count, using different criteria. Of course, I chose the higher one for this note. Further, the significance of the gene count is unclear. We now understand that many proteins can be made from a "single" gene (for example, by alternative splicing). Nevertheless, this stands, at least for now: the most genes known in any microbe, in fact, in any organism. Glossary entry: Alternative splicing.
News story in Microbe, 4/07: "Peculiar" T. vaginalis parasites are jam-packed with genes. forms.asm.org/microbe/index.asp?bid=49481.
Scientists Crack the Genome of the Parasite Causing Trichomoniasis. The press release from the New York Univ School of Medicine, one of the lead institutions for this work. January 11, 2007. http://communications.med.nyu.edu/ne...trichomoniasis.
The report of the Trichomonas genome: J M Carlton et al, Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207, 1/12/07. Online: http://www.sciencemag.org/content/315/5809/207.short.
Bacteria with "atypical" chromosomes
In the early days, it was very difficult to observe bacterial chromosomes. Bacteria are small, and their chromosomes are quite tiny by comparison with eukaryotic chromosomes. Further, bacterial chromosomes do not condense into more compact and more easily visible bodies, again in contrast to eukaryotic chromosomes. So, information about bacterial chromosomes emerged slowly, with various -- and mostly indirect -- techniques.
The early work suggested that bacteria have only one chromosome, and that it is circular. In one case, one E coli chromosome was even observed -- a circle. Other work seemed consistent with this, so the generality emerged: bacteria have only one chromosome, and it is circular. Both of these features served to distinguish bacteria from the eukaryotes. And that was nice: bacteria are supposedly "simpler", and having only one chromosome is certainly "simpler". Further, having a circular chromosome avoided the difficulties of replicating linear DNA, and could also be considered "simpler".
Alas, it is not really so. Neither feature is universal among bacteria. This web site discusses bacterial chromosomes, and has a table showing the number and type of chromosomes found in many bacteria. http://www.sci.sdsu.edu/~smaloy/MicrobialGenetics/topics/chroms-genes-prots/chromosomes.html. From Stanley Maloy, San Diego State University. It is part of a larger site on the broader topic of microbial genetics: http://www.sci.sdsu.edu/~smaloy/MicrobialGenetics/.
The following site is from a discussion of bacterial genetics in an online microbiology textbook. I include it here particularly because it contains a nice copy of a very famous figure, which I referred to above. Go to http://www.ncbi.nlm.nih.gov/books/NBK7908/; scroll down to Fig 5.2. This shows a single chromosome of one E coli cell, in the act of replicating. It is clearly circular.
That site is from Chapter 5, Genetics, by R K Holmes & M G Jobling, of the online book Medical Microbiology, 4th edition, edited by S Baron. This online book is listed in the Microbiology: books section of my page of Internet resources: Biology - Miscellaneous. The figure is from John Cairns, Cold Spring Harbor Symposia on Quantitative Biology 28:44, 1963.
Bacteria that can count -- and talk
Some bacteria can emit light -- more or less as fireflies do. The phenomenon is called bioluminescence. But the light from a single bacterial cell would be too dim to be of any use. So, isolated bacteria do not emit light. They only emit light when there are many of them together, so that -- together -- they give off a substantial amount of light. Clearly, bacteria can count how many neighbors they have.
How do bacteria count their population size? The basic logic of how they do it is actually rather simple. To make light, they need an "inducer" -- a substance that turns on the light-producing system. They make an inducer, and secrete it into the external environment. They then take it up from the environment. What does this accomplish? Well, imagine a simple situation of bacteria growing in a test tube. If there are only a few bacteria, there will be little inducer in the tube. When the bacteria try to take up inducer from the environment, they find very little -- and thus they do not emit light. But if the bacteria grow, so that there are many many bacteria in the tube, all making and secreting inducer, then the bacteria find a high level of inducer in the environment; they take it up, and emit light. Thus the bacteria sense their population size by responding to the level of inducer in the medium; as a result, they emit light only when the population size is large.
Is that artificial situation of a test tube of bacteria relevant to the bacteria in nature? Indeed it is. Some fish cultivate these bacteria in a special pouch, called a light organ. The light organ emits light only when it contains enough bacteria to do so usefully.
The phenomenon discussed above is often called quorum sensing. That is, the bacteria check to see if a quorum is present before emitting light. The details of this are now quite well understood. And as the system was being studied, it became clear that it was simply one example of a much wider phenomenon: bacteria communicating to each other, for a range of purposes. In this case, the bacteria are communicating their population size to their own kind. But more broadly, bacteria are signaling their presence -- and numbers -- to other types of bacteria too.
How hot?
What is the highest temperature at which life is possible? We all know that many of the molecules in living systems are quite sensitive to heat; the ease of cooking an egg reminds us of that regularly.
When I was in college, the highest temperature reported for life was around 60° C (degrees Celsius), the maximum temperature (Tmax) for growth of the bacterium Bacillus stearothermophilus. Since then, the known maximum has increased to at least 113° C, and perhaps even to 121° C. This increase came along with the discoveries of an entirely new class of microorganism, the archaea, and of a new geological phenomenon, the deep sea thermal vent; both discoveries date from 1977. Thus the increase in known Tmax for life is not simply an abstract story of some biological limit, but is part of a broad series of major advances in both biology and geology.
Thermus aquaticus has a maximum growth temperature of about 80° C. It was isolated from hot springs in Yellowstone National Park, and was reported in 1969. Thermus aquaticus is perhaps the organism that ushered in the new era of the commercialization of enzymes from thermopiles -- useful precisely because of their heat stability; the "Taq" DNA polymerase made the polymerase chain reaction -- PCR -- practical.
As noted above, 1977 brought the separate discoveries of archaea and deep sea thermal vents. Over the following years, these stories converged, and a succession of hyperthermophilic archaea were discovered near the vents. 1997 brought Pyrolobus fumarii, which grows up to 113° C; this archaeon has been widely accepted as having the highest known Tmax. 2003 brought a report of an archaeon that could grow at 121° C -- the normal operating temperature of an autoclave commonly used to kill even the most resistant forms of life, or so we thought. This organism has been dubbed simply Strain 121 for now.
These continuing discoveries of organisms with ever higher Tmax, maybe even up to the common operating temperature of an autoclave, raise some questions: ... [more]
Derek Lovley's web page (Univ Massachusetts) on the work that led to Strain 121: www.geobacter.org/Life-Extreme.
The first report of Strain 121. K Kashefi & D R Lovley, Extending the upper temperature limit for life. Science 301:934, 8/15/03. Online at http://www.sciencemag.org/content/301/5635/934.full.
For more about Lovley's lab, see "Electricigenic bacteria" in either the Redox section of the page for Internet resources for Intro Chem or the Carbohydrates section of the page for Internet resources for Intro Organic/Biochem.
For an update, see a summary of the Ninth International Conference on Thermophiles (Bergen, Norway, September 2007). T Satyanarayana, Meeting report: Thermophiles 2007. Current Science 93(10):1340, 11/25/07. Current Science, published by the Indian Academy of Sciences is freely available online. This article is at: http://www.ias.ac.in/currsci/nov252007/1340.pdf . The article contains a number of interesting tidbits. They suggest that the finding that strain S121 grows at 121° C has been questioned; they also suggest that another microbe has been shown to grow at 122° C at high pressure. It is normal enough that such claims are questioned. Time will tell. None of these details change the general perspective on high temperature microbes presented here.
Bacteria that know where north is
A microbiologist looks at a sample under the microscope. He notices that the bacteria seem to be moving over to one side of the microscope slide. Why? Perhaps they are responding to the light. So he adjusts the lighting, and it has no effect. After numerous such observations and tests, the conclusion is inescapable: the bacteria go north. Now, that is novel! He looks at the bacteria further, and finds that they contain tiny magnets -- iron oxide magnets, just like simple toy magnets. And that is how magnetic bacteria were discovered -- by Richard Blakemore in 1975.
Why do these bacteria use a magnet to guide their swimming? A common idea -- not entirely accepted -- is that these bacteria benefit from following the earth's magnetic lines of force. Doing that leads them "down" into the mud, which seems good for their lifestyle. Consistent with this, it was soon found that -- for some types of magnetic bacteria -- those in the northern hemisphere swim north, whereas those in the southern hemisphere swim south.
Magnetic Microbes, by Sandi Clement. commtechlab.msu.edu/Sites/dlc.../caOc96SC.html.
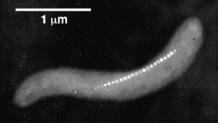
Magnetosomes. The figure at the right is from Richard Frankel's page: Magnetotactic Bacteria Photo Gallery. www.calpoly.edu/~rfrankel/mtbphoto.html. The figure shows a single cell of the bacterium Magnetospirillum magnetotacticum, with a chain of magnetosomes. Each individual magnetosome in the chain is approximately 45 nm across, and surrounded by a membrane. The Gallery page listed has many more figures, showing the diversity of magnetic bacteria. And for more, go to Frankel's home page, at Cal Poly San Luis Obispo: www.calpoly.edu/~rfrankel/. Scroll down to Research Interests, then Magnetotactic Bacteria.
R B Frankel & D A Bazylinski, Magnetosome mysteries. ASM News 70:176, 4/04. The news magazine ASM News -- now called Microbe -- is free online; this item is at forms.asm.org/microbe/index.asp?bid=26445.
C N Keim et al, Magnetoglobus, Magnetic aggregates in anaerobic environments. Microbe 2:437, 9/07. Microbe, the news magazine of the American Society for Microbiology is free online; this item is at forms.asm.org/microbe/index.asp?bid=52638. An article about a type of magnetic bacterium that normally occurs in multicellular aggregates. Should this be considered a multicellular bacterial organism? Considering that question gives insight into what multicellularity is about. This article is also listed in the section Multicellular bacteria.
W Hansen, This End Up -- Magnetic organelles point bacteria in the right direction. Berkeley Science Review, Issue 14, Spring 2008, p 8. A brief introduction to work on magnetic bacteria being done by Arash Komeili at UCB. Berkeley Science Review (BSR), published by UCB graduate students, is free online; this item is at sciencereview.berkeley.edu/ar...ticle=briefs_1.
The first report of magnetic bacteria: R Blakemore, Magnetotactic bacteria. Science 190:377-379, 10/24/75. The abstract is freely available at http://www.sciencemag.org/content/190/4212/377.abstract. You may or may not be able to get the full article at that site. If not and you have an institutional subscription to JStor, such as at UCB, try Access to Blakemore article through JStor.
Bacteria that eat other bacteria
The story of predatory bacteria starts with Bdellovibrio, a type of bacterium that obligatory lives within other bacterial cells. Since they kill the bacteria that they infect, Bdellovibrios form clear regions on a lawn of dense bacterial growth, much like bacterial viruses form plaques. But they are not viruses. They are cellular, with rather ordinary bacterial cells. It's just that they grow in a way that we find unusual. Well, it's not the "way" that is unusual as much as it is the "where". They burrow into a bacterial cell, and grow there.
E Jurkevitch, Predatory behaviors in bacteria -- diversity and transitions. Microbe 2:67, 2/07. Microbe, the news magazine of the American Society for Microbiology, is free online; this item is at forms.asm.org/microbe/index.asp?bid=48203.
At the end of the article listed above, Jurkevitch raises an interesting speculation about the possible role of predatory bacteria in the origin of the eukaryotic cell. Biologists agree that the mitochondrion arose from a bacterium that got inside another cell. But how did it get there? Bacteria do not show phagocytosis -- do not engulf other cells. However, predatory bacteria such as Bdellovibrio offer an alternative. Perhaps mitochondria originated by a predation event that led to symbiosis. There is no evidence on this point, so it must be regarded as speculation for now. At least it is a plausible view of how one of the great events of biological history might have occurred.
Multicellular bacteria
Single cells. Grow, and then divide into two. That is our simple image of bacteria. However, as we learn more about this vast group of organisms, we find that bacteria can be more complex. The myxobacteria probably have the most complex bacterial life cycle. They spend part of their life as free-living individual bacterial cells, then aggregate to form a fruiting body, an organized multicellular structure visible to the naked eye. In fact, their life cycle is rather similar to that of the cellular slime molds, such as Dictyostelium -- the myxomycetes.
Myxobacteria web page: http://myxobacteria.ahc.umn.edu/. In particular, step through the section What are the Myxobacteria? for a good introduction with some wonderful pictures. From Dr Martin Dworkin, with the help of Tim Leonard, at the University of Minnesota.
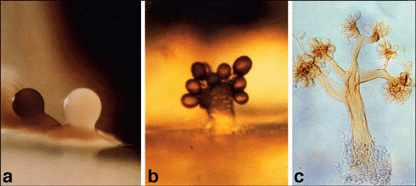 |
The figure above shows three examples of Myxobacteria fruiting bodies. Each is just under one millimeter tall. The figure is Fig 1 from the Microbe article listed below, and is also shown at the Myxobacteria web page listed above (What Are Myxos? : Part 2). The photos are attributed at both places to Dr Hans Reichenbach. |
M Dworkin, Lingering Puzzles about Myxobacteria. Microbe 2:18, 1/07. Dworkin's "puzzles" include:
- How the cells construct the multicellular, macroscopic fruiting body
- The biochemical basis of myxospore morphogenesis
- The mechanism and function of individual cellular motility
- The regulation of directionality of social movement
- The mechanism of the cells' ability to perceive physical objects at a distance
- The role of the myxobacteria in nature.
Microbe, the news magazine of the American Society for Microbiology, is free online; this item is at forms.asm.org/microbe/index.asp?bid=47794 (HTML) or forms.asm.org/ASM/files/ccLib...0107000018.pdf (PDF).
C N Keim et al, Magnetoglobus, Magnetic aggregates in anaerobic environments. Microbe 2:437, 9/07. Microbe, the news magazine of the American Society for Microbiology is free online; this item is at forms.asm.org/microbe/index.asp?bid=52638. An article about a type of magnetic bacterium that normally occurs in multicellular aggregates. Should this be considered a multicellular bacterial organism? Considering that question gives insight into what multicellularity is about. This article is also listed in the section Bacteria that know where north is. Other topics on this page introduce other ways in which some bacteria are more complex that we might have thought. These include:
A huge virus
Ordinary organisms are based on cells. The organisms reproduce by the cells growing and dividing. Viruses are different. Viruses are small and simple. Viruses do not grow and divide. They reproduce by infecting a cell, disassembling, and then directing the production of new "parts", which then assemble into new virus particles.
Small and simple? Well, usually. The smallest viruses have one millionth or so the amount of genome (DNA or RNA) we do. Some have only a handful of genes. Some have only a piece of DNA (or RNA) and a simple protein coat -- no machinery for making anything, and no enzymes.
Some viruses are not so small and not so simple. Biologists have still been able to make a clear distinction between viruses and cells, primarily by looking at their basic strategy for reproduction. Cells grow and divide; viruses disassemble and reassemble.
The most recent challenge to the simplicity of viruses is the mimivirus, which grows in the protozoan Acanthamoeba polyphaga. It has about three times more genetic material (DNA) than any previously known virus -- more DNA than some bacteria. It is bigger than some bacteria -- about 400 nm (0.4 μm) diameter. And it is quite complex, with a collection of enzymes that are supposedly not to be found in viruses. For example, mimivirus codes for several enzymes used in protein synthesis -- genes never before found in any virus. Yet its life style (and structure) make it clear that this is a virus. It was characterized as a virus only in 2003.
2008 brings new developments that make the story of mimivirus even more fascinating. First, a new mimivirus, even bigger than the first. They call it mamavirus. But perhaps more importantly, a satellite virus: a virus that can grow only in cells infected by mimivirus. A news story about this satellite virus, dubbed Sputnik: 'Sputnik' Virus Orbits, Hijacks Other Viruses, Aug. 13, 2008. dsc.discovery.com/news/2008/0...s-sputnik.html.
Discussions about mimivirus and Sputnik inevitably seem to wander onto topics such as "What is a virus?" or even "What is life?" These are fun to discuss, but a caution: they need not have simple answers, or even any answers at all beyond our common definitions. Use such questions to provide a framework for your knowledge and understanding, but forcing simple answers to complex questions is not fruitful. Mimivirus has its own website: http://www.giantvirus.org.
The organism now known as mimvirus was found in 1992. The first paper that identified it as a virus: B La Scola et al, A giant virus in Amoebae. Science 299:2033, 3/28/03. Online: http://www.sciencemag.org/content/299/5615/2033.short.
The report of the Sputnik satellite virus: B La Scola et al, The virophage as a unique parasite of the giant mimivirus. Nature 455:100, 9/4/08. There is a good news story about this finding: Biggest known virus yields first-ever virophage. Microbe 3:505, 11/08. Free online: microbemagazine.org/images/st...1108000502.pdf. Scroll down to the story, on page 4 of the file.
The biggest microbe?
Probably the unicellular green alga Acetabularia, whose cells can be several centimeters long. Because of the large size, Acetabularia was a favorite organism for studying the relationship between nucleus and cytoplasm. The following links introduce the organism and some classic experimental work.
- en.Wikipedia.org/wiki/Acetabularia. Basic introduction to Acetabularia.
- www.accessexcellence.org/RC/V...mmerling_s.php. Classic work on the role of nucleus and cytoplasm in determining cell development, done by J Hammerling in the 1930s. The large cells of Acetabularia allowed a simple but novel transplantation to be done; the results revealed the key role of the nucleus. This item is given as a link at the end of the previous one. (Another Access Excellence page is listed on this page, in the section Big bacteria. The Access Excellence site is listed as a general resource on my page of Miscellaneous Internet Resources, under Of local interest... -- since it had its origins near here.)
Briefly noted
This section is something of a "miscellany" -- a place to briefly note some other unusual aspects of microbial life. In some cases, I may make only a single small point or note only a single paper, Perhaps some of these will grow into "full-blown" topics at some point, or perhaps we will just keep a section of "miscellany".
G forces? Humans don't do well with g forces a few times normal gravity. Microbes do better, it seems. A recent paper shows that several microbes studied, bacteria and yeast, grew in an ultracentrifuge tube with accelerations many thousands of times g. Two of the bacterial grew at the highest accelerations tested, over 400,000 x g. They have no information about what limits the growth as the g force increases; they speculate that it has something to do with sedimentation within the cell. That organisms vary might allow them to pursue finding what is important. It is also unclear why this is of interest. After all, such high g forces are found in nature only under extreme conditions, such as the shock waves of supernovae. For now, this paper is basically just a cute finding. It will be interesting to see where it leads.
- News story... Bacteria Grow Under 400,000 Times Earth's Gravity (National Geographic, April 25, 2011): http://news.nationalgeographic.com/news/2011/04/110425-gravity-extreme-bacteria-e-coli-alien-life-space-science/.
- The paper... S Deguchi et al, Microbial growth at hyperaccelerations up to 403,627 x g. PNAS 108:7997, May 10, 2011. Online at: http://www.pnas.org/content/108/19/7997.
Where is the inside? Some bacteria, such as the gram negatives, have a double membrane system. It is the inner membrane that is energized, and used to make ATP. Now we have a discovery of the first double membrane system of an archaeon -- and it is the outer membrane that is energized. The archaeon, Ignicoccus hospitalis, is closely associated with Nanoarchaeum equitans -- which relies on the Ignicoccus for its energy; is this energy parasitism dependent on the unusual energy system of the Ignicoccus? The authors even wonder whether Ignicoccus might be an ancestor of the eukaryotic cell. Clearly, this is an unusual and intriguing finding -- still quite incomplete.
For a fine introduction to this novel system, see the ASM blog entry by Moselio Schaechter... Of Archaeal Periplasm & Iconoclasm (February 11, 2010): http://schaechter.asmblog.org/schaechter/2010/02/of-archaeal-periplasm-iconoclasm.html.
The paper... U Küper et al, Energized outer membrane and spatial separation of metabolic processes in the hyperthermophilic Archaeon Ignicoccus hospitalis. PNAS 107:3152, 2/16/10. Online at: http://www.pnas.org/content/107/7/3152.
Arsenic. There are bacteria that can oxidize arsenic compounds, and there are bacteria that can reduce arsenic compounds. Now there is a report of bacteria that can use arsenite -- AsO33-, containing As(III) -- as the electron donor for photosynthesis. (The most common electron donor is water -- with oxygen gas being evolved. The most common electron donor in anaerobic systems is sulfide, often with sulfur granules being produced.) Analysis of this process suggests that arsenic metabolism is quite ancient, and that it is an important part of the arsenic cycle in nature. News story: In Lake, Photosynthesis Relies on Arsenic, August 18, 2008. http://www.nytimes.com/2008/08/19/science/19obarsenic.html.
The paper... T R Kulp et al, Arsenic(III) Fuels Anoxygenic Photosynthesis in Hot Spring Biofilms from Mono Lake, California. Science 321:967, 8/15/08. Online at: http://www.sciencemag.org/content/321/5891/967.abstract.
Microbes survive the cold. Scientists have recovered DNA and even viable bacteria from ancient ice samples in the Antarctic (and other places). The idea is that bacteria were trapped in the ice, perhaps in pockets of liquid water just big enough for the one cell. The bacteria may have carried out maintenance reactions, perhaps only a few chemical reactions per day, to survive. Even with quibbling about how old each sample really is, this is still a fascinating insight into survival of life in extreme conditions. News story: Eight-million-year-old bug is alive and growing, August 7, 2007. http://www.newscientist.com/article/dn12433.
Here are a couple of papers, both of which should be freely available. The first goes with the news story listed above, and is generally about the isolation of the old bacteria and their DNA. The second, from UC Berkeley, is about how the bacteria may metabolize and survive in the ice.
K D Bidle et al, Fossil genes and microbes in the oldest ice on Earth. PNAS 104:13455, 8/14/07. Free online at: http://www.pnas.org/content/104/33/13455.abstract.
R A Rohde & P B Price, Diffusion-controlled metabolism for long-term survival of single isolated microorganisms trapped within ice crystals. PNAS 104:16592, 10/16/07. Free online at: http://www.pnas.org/content/104/42/16592.abstract.
A lonely bug. Organisms live in complex communities. Seems pretty basic in our modern understanding of biology. Certainly, we expect to find bacteria in complex communities. So, it is striking when we find a report of the discovery of a bacterial growth in a South African goldmine that seems to contain only one species. Of course, it is hard to exclude some very low level of other organisms, but the analysis shows that the main bacterium, called Candidatus Desulforudis audaxviator, is at least 99.9% of the culture.
What is it growing on down there? Well, seems likely that it is using the energy from uranium decay as its main energy source. So this loner is also a nuclear-powered bug.
The paper is: D. Chivian et al, Environmental Genomics Reveals a Single-Species Ecosystem Deep Within Earth . Science 322:275, 10/10/08. Free online at: http://www.sciencemag.org/content/322/5899/275.abstract. For a good news story about this work, see Journey Toward The Center Of The Earth: One-of-a-kind Microorganism Lives All Alone, 10/10/08: http://http://www.sciencedaily.com/releases/2008/10/081009143708.htm.
More "Curious microbes"
While looking for some nice web sites to include in the various sections above, I came across Sandi Clement's page on Magnetic Microbes listed for Bacteria that know where north is. Turns out that is part of a larger site with a theme rather similar to this one -- and written by students in a class on Extreme and Unusual Microbes taught by Dr. Rick Martin at the Center for Microbial Ecology, Michigan State Univ. The site is called The Curious Microbe - Essays of the Extreme and the Unusual: commtechlab.msu.edu/Sites/dlc...us/cindex.html.
Contributors
- Robert Bruner (http://bbruner.org)
This page viewed 15659 times
The BioWiki has 63323 Modules.

