7.3: The "One Gene- One Enzyme" Hypothesis
- Page ID
- 132193
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)7.3 The "One Gene: One Enzyme" Hypothesis
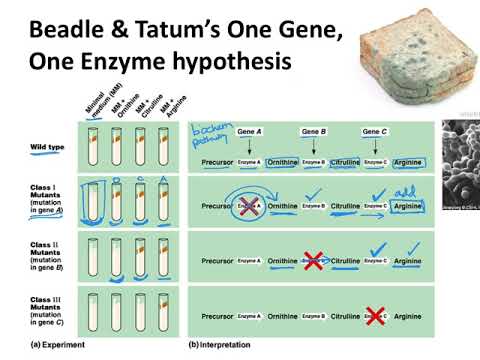
Beadle and Tatum’s experiments are important not only for their conceptual advances in understanding genes, but also because they demonstrate the utility of screening for genetic mutants to investigate a biological process — this is called genetic analysis.
Beadle and Tatum’s results were useful to investigate biological processes, specifically the metabolic pathways that produce amino acids. For example, Srb and Horowitz (1944) tested the ability of the amino acids to rescue auxotrophic strains. They added one of each of the amino acids to minimal medium and recorded which of these restored growth to independent mutants.
Watch the video below, BIOL 183: Beadle & Tatum’s One-Gene-One-Enzyme hypothesis, by Susan Bush (2020) at Metropolitan State University on YouTube, which explains the one – gene – one – enzyme hypothesis.
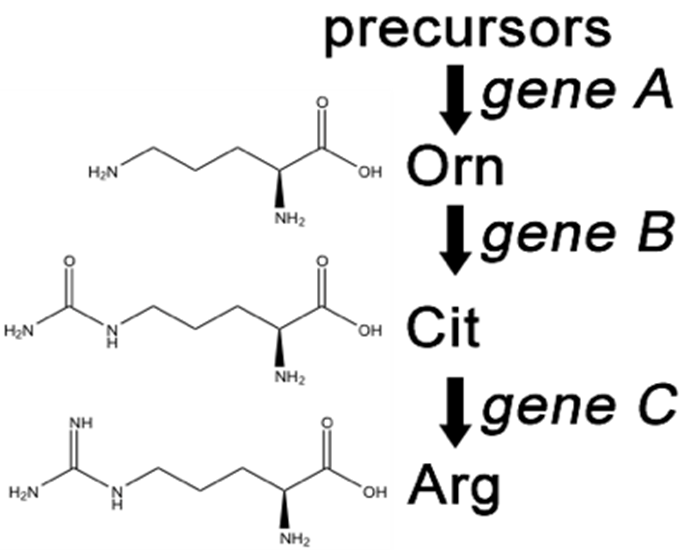
A convenient example is arginine. If the progeny of a mutagenized spore could grow on minimal medium only when it was supplemented with arginine (Arg), then the auxotroph must bear a mutation in the Arg biosynthetic pathway and was called an “arginineless” strain (arg-).
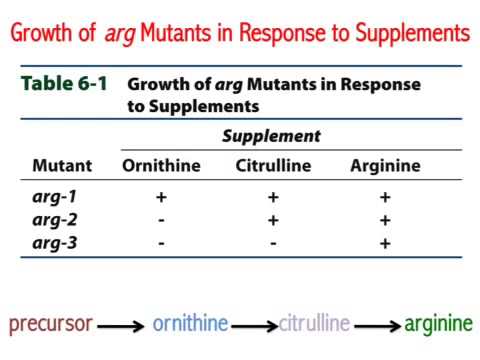
Synthesis of even a relatively simple molecule, such as arginine, requires many steps — each with a different enzyme. Each enzyme works sequentially on a different intermediate in the pathway (Figure 7.3.1). For arginine (Arg), two biochemical intermediates are ornithine (Orn) and citrulline (Cit). Thus, mutation of any one of the enzymes in this pathway could turn Neurospora into an Arg auxotroph (arg-). Srb and Horowitz extended their analysis of Arg auxotrophs by testing the intermediates of amino acid biosynthesis for the ability to restore growth of the mutants (Figure 7.3.2).
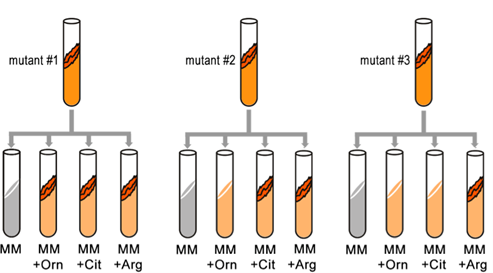
They found that only Arg could rescue all the Arg auxotrophs, while either Arg or Cit could rescue some (Table 7.3.1). Based on these results, they deduced the location of each mutation in the Arg biochemical pathway (i.e., which gene was responsible for the metabolism of which intermediate).
| Mutants In | MM + Orn | MM + Cit | MM + Arg |
|---|---|---|---|
| gene A | Yes | Yes | Yes |
| gene B | No | Yes | Yes |
| gene C | No | No | Yes |
The video below, Gene Interactions P1, by Michelle Stieber (2014) on YouTube, discusses gene interactions and related biochemical pathways.
Media Attributions
- Figure 7.3.1Original by Deyholos (2017), CC BY-NC 3.0, Open Genetics Lectures
- Figure 7.3.2Original by Deyholos (2017), CC BY-NC 3.0, Open Genetics Lectures
References
Bush, S. (2020, April 16). BIOL 183: Beadle & Tatum’s one-gene-one-enzyme hypothesis (video file). YouTube. https://www.youtube.com/watch?v=4nXX2djQVvI
Deyholos, M. (2017). Figures: 4. A simplified version of the Arg biosynthetic pathway… and 5. Testing different Arg auxotrophs for their ability to grow…(digital image). In Locke, J., Harrington, M., Canham, L. and Min Ku Kang (Eds.), Open Genetics Lectures, Fall 2017 (Chapter 3, p. 3). Dataverse/ BCcampus. http://solr.bccampus.ca:8001/bcc/file/7a7b00f9-fb56-4c49-81a9-cfa3ad80e6d8/1/OpenGeneticsLectures_Fall2017.pdf
Srb, A. M. & Horowitz N. H. (1944). The ornithine cycle in Neurospora and its genetic control. Journal of Biological Chemistry, 154, 129-139. https://doi.org/10.1016/S0021-9258(18)71951-0
Stieber, M. (2014, April 12). Gene interactions P1 (video file). YouTube. https://www.youtube.com/watch?v=Fv7UtsPfF-A
Long Descriptions
- Figure 7.3.1 The arginine biosynthetic pathway, showing the organic structures of citrulline and ornithine as intermediates in arginine metabolism. These chemical reactions depend on enzymes, which are the products of three different genes: A, B and C. [Back to Figure 7.3.1]
- Figure 7.3.2 Three mutants are tested for their ability to grow on minimal media, or minimal media supplemented with ornithine, citrulline, or arginine. Depending on the gene that is not functional in the particular mutant, they may or may not thrive in various media, therefore only arginine could rescue all the arginine auxotrophs, while either ornithine or citrulline could rescue others. [Back to Figure 7.3.2]