4.3: Modes of Inheritance
- Page ID
- 132164
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)4.3 Modes of Inheritance
Usually, we are presented with a pedigree of an uncharacterized disease or trait, and one of the first tasks is to determine which modes of inheritance are possible, and then, which mode of inheritance is most likely. This information is essential in calculating the probability that the trait will be inherited in any future offspring. We will mostly consider five major types of inheritance: autosomal dominant (AD), autosomal recessive (AR), X-linked dominant (XD), X-linked recessive (XR), and Y-linked (Y) inheritance.
We generally make two assumptions in analyzing Pedigree Charts. These are as follows:
- Complete Penetrance — an individual in the pedigree will be affected (express the phenotype associated with a trait) when the individual carries at least one dominant allele of a dominant trait, or two recessive alleles of a recessive a trait.
- Rare-in-Population — generally, the trait in question is rare in the general population.
The following are some hints and clues to help us interpret Pedigree Charts:
- An unaffected individual cannot have any alleles of a dominant trait (because a single allele of a dominant trait causes an individual to be affected).
- Individuals marrying into the family are assumed to have no disease alleles – they will never be affected and can never be carriers of a recessive trait (because the trait is rare in the population).
- An unaffected individual can be a carrier (have one allele) of a recessive trait (because two alleles of a recessive trait are required for an individual to be affected).
- When a trait is X-linked, a single recessive allele is sufficient for a male to be affected (because the male is hemizygous – he only has one allele of an X-linked trait).
- A father transmits his allele of X-linked genes to his daughters, but not his sons. A mother transmits an allele of X-linked genes to both her daughters and her sons.
Take a look at the following video, Pedigree Analysis, by AK Lecture Series (2015) on YouTube, which discusses Pedigree Charts and how to analyze them.
Let us now take a look at the various modes of inheritance and typical pedigree charts which are characteristic of each mode.
Autosomal Dominant (AD)
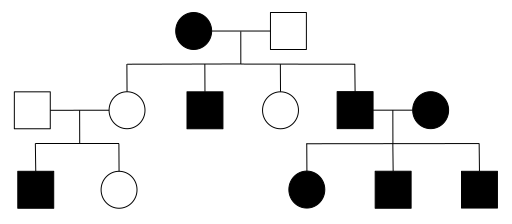
When a disease is caused by a dominant allele of a gene, every person with that allele will show symptoms of the disease (assuming complete penetrance), and only one disease allele needs to be inherited for an individual to be affected. Thus, every affected individual must have an affected parent. A pedigree with affected individuals in every generation is typical of AD diseases. However, beware that other modes of inheritance can also show the disease in every generation, as described below. It is also possible for an affected individual with an AD disease to have a family without any affected children, if the affected parent is a heterozygote. This is particularly true in small families, where the probability of every child inheriting the normal, rather than disease allele is not extremely small. Note that AD diseases are usually rare in populations, therefore affected individuals with AD diseases tend to be heterozygotes (otherwise, both parents would have had to been affected with the same rare disease). Huntington Disease, Achondroplastic dwarfism, and Polydactyly are all examples of human conditions that may follow an AD mode of inheritance.

Example: Achondroplasia is a common form of dwarfism. FGFR3 gene at 4p16 (chromosome 4, p arm, region 1, band 6) encodes a receptor protein that negatively regulates bone development. A specific base pair substitution in the gene makes an over-active protein and this results in shortened bones. Achondroplasia is considered autosomal dominant because the defective proteins made in A / a embryos halt bone growth prematurely. A / A embryos do not make enough limb bones to survive. Most, but not all dominant mutations are also recessive lethal. In achondroplasia, the A allele shows dominant visible phenotype (shortness) and recessive lethal phenotype.
X-Linked Dominant (XD)
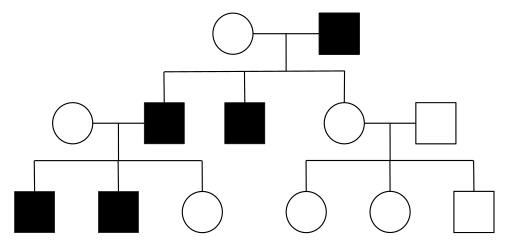
In X-linked dominant inheritance, the gene responsible for the disease is located on the X-chromosome, and the allele that causes the disease is dominant to the normal allele in females. Because females have twice as many X-chromosomes as males, females tend to be more frequently affected than males in the population. However, not all pedigrees provide sufficient information to distinguish XD and AD. One definitive indication that a trait is inherited as AD, and not XD, is that an affected father passes the disease to a son; this type of transmission is not possible with XD, since males inherit their X chromosome from their mothers.

Example: fragile x syndrome — The FMR1 gene at Xq21 (X chromosome, q arm, region 2, band 1) encodes a protein needed for neuron development. There is a (CGG)n repeat array in the 5’UTR (untranslated region). If there is expansion of the repeat in the germline cell the child will inherit a non-functional allele. XA / Y males have fragile X mental retardation (IQ < 50) because none of their neurons can make FMR1 proteins. Fragile X syndrome is considered X-linked dominant because only some neurons in XA / Xa females can make FMR1 proteins. The severity (IQ 50 – 70) in these females depends upon the number and location of these cells within in the brain.
Autosomal Recessive (AR)
Diseases that are inherited in an autosomal recessive pattern require that both parents of an affected individual carry at least one copy of the disease allele. With AR traits, many individuals in a pedigree can be carriers, probably without knowing it. Compared to pedigrees of dominant traits, AR pedigrees tend to show fewer affected individuals and are more likely than AD or XD to “skip a generation”. Thus, the major feature that distinguishes AR from AD or XD is that unaffected individuals can have affected offspring. Attached earlobes is a human condition that may follow an AR mode of inheritance.
AR example: phenylketonuria (PKU) – Individuals with phenylketonuria (PKU) have a mutation in the PAH gene at 12q24 (chromosome 12, q arm, region 2, band 4), which encodes an enzyme that breaks down phenylalanine into tyrosine called phenylalanine hydrolase (PAH). Without PAH, the accumulation of phenylalanine and other metabolites, such as phenylpyruvic acid, disrupts brain development, typically within a year after birth, and can lead to intellectual disability. Fortunately, this condition is both easy to diagnose and can be successfully treated with a low phenylalanine diet. There are over 450 different mutant alleles of the PAH gene, so most people with PKU are compound heterozygotes . Compound heterozygotes have two different mutant alleles (different base pair changes) at a given locus, in this case the PAH gene.
X-Linked Recessive (XR)
Because males have only one X-chromosome, any male that inherits an X-linked recessive disease allele will be affected by it (assuming complete penetrance). Therefore, in XR modes of inheritance, males tend to be affected more frequently than females in a population. This contrasts with AR and AD, where both sexes tend to be affected equally, and XD, in which females are affected more frequently. Note, however, in the small sample sizes typical of human families, it is usually not possible to accurately determine whether one sex is affected more frequently than others. On the other hand, one feature of a pedigree that can be used to definitively establish that an inheritance pattern is not XR is the presence of an affected daughter from unaffected parents; because she would have had to inherit one X-chromosome from her father, he would also have been affected in XR.
XR example: hemophilia A- F8 gene at Xq28 (X chromosome, q arm, region 2, band 8) encodes blood clotting factor VIIIc. Without Factor VIIIc, internal and external bleeding can’t be stopped. Back in the 1900s, Xa / Y male’s average life expectancy was 1.4 years, but in the 2000s it has increased to 65 years with the advent of Recombinant Human Factor VIIIc. Hemophilia A is recessive because XA / Xa females have normal blood coagulation, while Xa / Xa females have hemophilia.
Y-Linked
Only males are affected in human Y-linked inheritance (and other species with the X/Y sex determining system). There is only father-to-son transmission. This is the easiest mode of inheritance to identify, but it is one of the rarest because there are so few genes located only on the Y-chromosome.
A common, but incorrect, example of Y-linked inheritance is the hairy-ear-rim phenotype seen in some Indian families. A better example are the Y-chromosome DNA polymorphisms that have been used to follow the male lineage in large families or through ancient ancestral lineages. For example, the Y-chromosome of Mongolian ruler Genghis Khan (1162-1227 CE), and his male relatives, accounts for ~8% of the Y-chromosome lineage of men in Asia (0.5% world wide).
Media Attributions
- Figure 4.3.1 Autosomal dominant by Simon Caulton, CC BY-SA 3.0, via Wikimedia Commons
- Figure 4.3.2 Wiki Drawing – X-Linked Dominant (1) by Madibc68, CC BY-SA 4.0, via Wikimedia Commons
- Figure 4.3.3 Autosomal Recessive Pedigree Chart by Jerome Walker, CC BY-SA 3.0, via Wikimedia Commons
- Figure 4.3.4 Wiki Drawing – X-Linked Recessive (1) by Madibc68, CC BY-SA 4.0, via Wikimedia Commons
- Figure 4.3.5 Wiki Drawing – Y-Linked (1) by Madibc68, CC BY-SA 4.0, via Wikimedia Commons
Reference
AK Lecture Series. (2015, January 13). Pedigree Analysis (video file). YouTube. https://youtu.be/Wgmgt_Ph6Ko
Long Descriptions
- Figure 4.3.1 Pedigree chart showing inheritance of an autosomal dominant trait over four generations. Affected females are shown as red-coloured circles; normal females are blue-coloured circles; affected males are red-coloured squares; normal males are blue-coloured squares. Generation I begins with a normal male and an affected female, mating to produce five offspring. Their offspring are: two affected and three unaffected. Two of these Generation II individuals mate, and their progeny is shown, along with a final Generation IV, with the characteristic pattern for autosomal dominant traits depicted. [Back to Figure 4.3.1]
- Figure 4.3.2 Pedigree chart showing the inheritance of a X-linked dominant trait over three generations. An affected female mates with a normal male in Generation I, to produce four offspring – one normal male, one affected male, one normal female and one affected female. The normal female mates with a normal male, and they produce two unaffected children. The affected male of Generation II mates with a normal female, and they produce four children – two affected females and two unaffected males. This pattern is characteristic for X-linked dominant inheritance. [Back to Figure 4.3.2]
- Figure 4.3.3 Pedigree chart showing the inheritance of an autosomal recessive trait, over three generations. Red and blue colours depict differences in the male and female. Both colours mean heterozygous, solid red colour means homozygous recessive, and solid blue color means wild type or homozygous dominant. Generation I begins with two carrier parents, who are therefore heterozygous. They produce four children, two are normal, one is a carrier and one is affected. This pattern is consistent with the inheritance of autosomal recessive traits. [Back to Figure 4.3.3]
- Figure 4.3.4 Pedigree chart showing the inheritance of a X-linked recessive trait over three generations. Generation I outlines one affected female and one affected male, mating to produce four children, two unaffected females and two affected males. One of the unaffected females mates with a normal man, and they produce two children: one affected male and an unaffected female. This indicates the unaffected mother was a carrier. This pattern of inheritance is typical of an X-linked recessive trait. [Back to Figure 4.3.4]
- Figure 4.3.5 Pedigree chart showing the inheritance of a Y-linked trait. Three generations are shown, starting with a normal female and an affected male, who produce three offspring. Offspring are: one normal female and two affected males. One affected male mates with a normal female, and their offspring comprises one normal female and two affected males. This pattern of inheritance is typical of Y-linked traits. [Back to Figure 4.3.5]