33.3: Genetic Epidemiology
- Page ID
- 41317
Genetic epidemiology focuses on the genetic factors contributing to disease. Genome-Wide association studies (GWAS), previously described in depth, identify genetic variants that are associated with a particular disease while ignoring everything else that may be a factor. With the decrease of whole genome sequencing, these types of studies are becoming much more frequent.
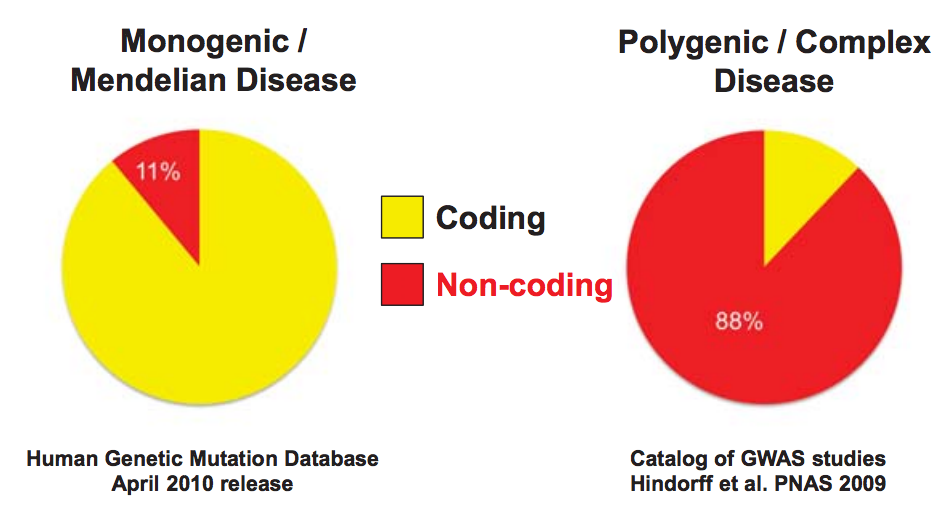
source unknown. All rights reserved. This content is excluded from our Creative
Commons license. For more information, see http://ocw.mit.edu/help/faq-fair-use/.
In genetic epidemiology there are many genetic factors that you can test to identify diseases in a particular individual. You can look at family risk alleles which are inherited with a common trait in specific genes or variants. You can study monogenic, actionable protein-coding mutations which are the most understood, would have the highest impact, and would be the easiest to interpret. There is the possibility of testing all coding SNPs (single nucleotide polymorphisms) with a known disease association. There are debates over whether a patient needs to or would want to know this information sometimes especially if the disease is not treatable. A person’s quality of life may decrease just from knowing they may have the untreatable disease even if no symptoms are exhibited. You can also test all coding and non-coding associations from GWAS, all common SNPs regardless of association to any disease, or the whole genome.
Did You Know?
23andMe is a personal genomics company that offers saliva-based direct-to-consumer genome tests. 23andMe gives consumers raw genetic data, ancestry-related results, and estimates of predisposition for more than 90 traits and conditions. In 2010, the FDA notified several genetic testing companies, including 23andMe, that their genetic tests are considered medical devices and federal approval is required to market them. In 2013, the FDA ordered 23andMe to stop marketing its Saliva Collection Kit and Personal Genome Service (PGS) as 23andMe had not demonstrated that they have “analytically or clinically validated the PGS for its intended uses” and the “FDA is concerned about the public health consequences of inaccurate results from the PGS device” [? ]. The FDA expressed concerns over both false negative and false positive genetic risk results, saying that a false positive may cause consumers to undergo surgery, intensive screening, or chemoprevention in the case of BRCA-related risk, for example, while a false negative may prevent consumers from getting the care they need. In class, we discussed whether people should be informed about potential risk alleles they may carry. Often, people may misunderstand the probabilities provided to them and either underestimate or overestimate how concerned they should be. The argument was also raised that people should not be told they are at risk if there is nothing current medicine and technology can do to mitigate the risk. If people are going to be informed about a risk, the risk should be actionable; i.e. they should be able to do something about it, instead of just live in worry, as that added stress may cause other health problems for them.
Not only is there the choice of what to test, there is the question of when to test someone for a particular condition. Diagnostic testing occurs after symptoms are displayed in order to confirm a hypothesis or distinguish between different possibilities of having a condition. You can also test predictive risk which occurs before symptoms are even shown by a patient. You may test newborns in order to intervene early or even do pre-natal testing via an ultrasound, maternal serum, probes or chorionic villus sampling. In order to test which disorders you may pass on to your child, you can do pre-conception testing. You can also do carrier testing to determine if you are a carrier of a particular mutant allele that may run in your family history. Testing genetics and biomarkers can be tricky because it can be unknown if the genetics or biomarker seen is causing the disease or is a consequence of having the disease.
To interpret disease associations, we need to use epigenomics and functional genomics. The genetic associations are still only probabilistic: if you have a genetic variant, there is still a possibility that you will not get the disease. Based on Bayesian statistics however, the posterior probability increases if the prior increases. As we find more and more associations and variants, the predictive value will increase.

