24.2: Experimental Techniques
- Page ID
- 41091
The ENCODE project used a wide range of experimental techniques, ranging from RNA-seq, CAGE-seq, Exon Arrays, MAINE-seq, Chromatin ChIP-seq, DNase-seq, and many more.

ENCODE Project. All rights reserved. This content is excluded from our Creative
Commons license. For more information, see http://ocw.mit.edu/help/faq-fair-use/.
One of the most important techniques used was ChIP-seq (chromatin immunoprecipitation followed by sequencing). The first step in a ChIP experiment is to target DNA fragments associated with a specific protein. This is done by using an anti-body that targets the specific protein and is used to immunoprecipitate the DNA-protein complex. The final step is to assay the DNA. This will determine the sequences bound to the proteins.
ChIP-seq has several advantages over previous techniques (e.g. ChIP-chip). For example, ChIP-seq has single nucleotide resolution and its alignability increases with read length. However, ChIP-seq has several disadvantages. Sequencing errors tend to increase substantially near the end of reads. Also, with low number of reads, sensitivity and specificity tend to decrease when detecting enriched regions. Both of these problems arise when processing the data and many of the computational techniques seek to rectify this.
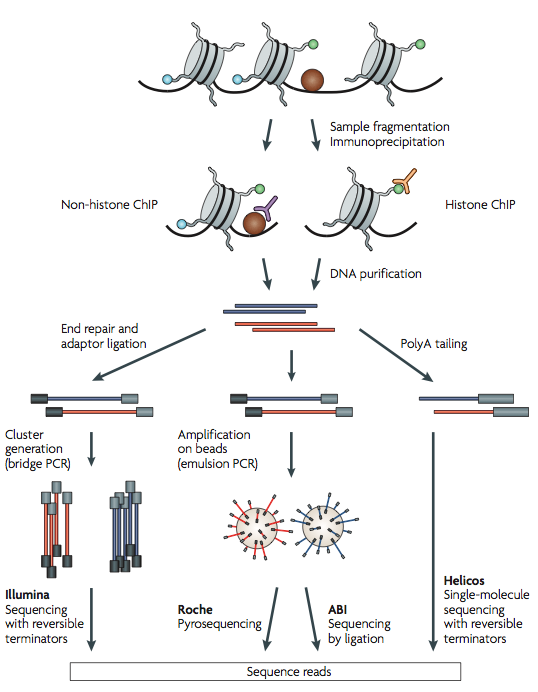
Courtesy of Macmillan Publishers Limited. Used with permission.
Source: Park, Peter J. "ChIP-seq: Advantages and Challenges of a
Maturing Technology." Nature Reviews Genetics 10, no. 10 (2009): 669-80

