14.4: Read Mapping - Spaced Seed Alignment
- Page ID
- 40998
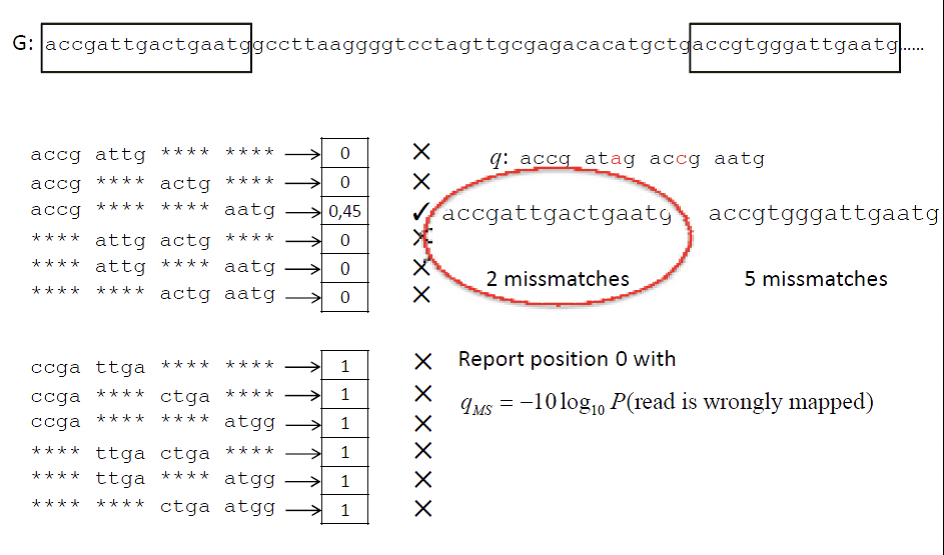
The idea behind read mapping is to align the sequenced reads to a reference genome. Sequence alignment algorithms discussed in earlier chapters will not work for this case due to the scale of the problem. The goal is to align millions of reads to the genome and would take too long if each was aligned individually. Instead, we will introduce the Spaced Seed Alignment approach. This process begins by using the reference genome to creating a hash table of 8-mers, which do not have to be contiguous. The positions of these stored spaced seeds are mapped to the hash table. Using these spaced 8-mers, each read is then compared with each possible position in the reference genome and scored based on the number of base pair matches (Figure 2).
More accurately, for each position, it is possible to calculate the score using the equation qMS = −10 log10(1 − P (i|G, q)), where P (i|G, q) represents the probability that the read, q, is mapped to posi- tion i of reference genome G. More details on deriving this score can be found in Figure 13.2.
It is possible to adjust the parameters of this method in order to alter the sensitivity, speed, and memory
of the algorithm. Using smaller k-mer seeds allows for less precise base pair matching (greater sensitivity), but requires more matches to be attempted. Smaller seeds take up less memory, while larger seeds run faster.
There exist methods other than the one described above to perform this alignment. The most popular of which is the Burrows-Wheeler approach. The Burrows-Wheeler transform is an even more efficient algorithm for mapping reads and will be discussed in a later chapter. It is able to speed up the process of finding matches in the large genome by reordering the genome in a very specific permutation. This allows reads to be matched solely as a function of the length of the read and not the genome. As better sequencing technology allows for larger read lengths, more algorithms will need to be developed to handle the extra processing.
Unlike ChIP-Seq, a similar technology, RNA-seq is more complex. This is because the read mapper needs to worry about small exons interspersed between large introns and be able to find both sides of an exon. This complexity can be overcome by using the above mentioned spaced seed matching technique, and detecting when two k-mers from the same read are separated by a long distance. This would signal a possible intron and can be fixe by then extending the k-mers to fill in gaps (SNO methods). Another method is to base the alignment on contiguous reads, which are further fragmented into 20-30 bp regions. These regions are remapped, and the positions with two or more different alignments are marked as splice junctions. Exon-first aligners are faster than the previous methods, but come at a cost: they fail to differentiate psuedogenes, prespliced genes, and transposed genes.


