12.7: Technologies- in the wet lab, how can we find these?
- Page ID
- 40991
How would we find ncRNAs? We have about 20-30 examples of ncRNAs with evidence of importance, but more are out there. Chromatin state maps (from ENCODE, chip-seq) can be used to find transcriptional units that do not overlap proteins. We can walk along map and look for genes (look by eye at chromatin map to find ncRNAs). Nearly 90% of time such a signature is found, RNA will be transcribed from it. We can validate this through northern blot
When looking at a chromatin map to find ncRNAs, we are essentially looking through the map with a window of a given size and seeing how much signal vs. noise we are getting, compared to what we might expect from a random-chance hypothesis. As both large and small windows have benefits, both should be used on each map section. Larger windows encapulate more information; smaller windows are more sensitive.
After finding integenic regions, we find conserved regions.
We check if new regions are under selective pressure; fewer mutations in conserved regions. If a nucleotide never has a mutation between species, it’s highly conserved.
linc-RNAs are more conserved than introns, but less conserved than protein-coding introns, possibly due to non-conserved sequences in loop regions of lincRNAs.
Finding what lincRNAs’ functions are: “Guilt by association”: We can find proteins that correlate with particular lincRNA in terms of expression; lincRNAs are probably correlated to a particular pathway. In this way, we acquire a multidimensional barcode for each lincRNA (what it is and is not related to). We Can cluster lincRNA signatures and identify common patterns. Lots have to do with cell cycle genes. (This approach works 60-70% of the time)
As most lincRNAs are over 3000 bases, many contain sequences for 100 amino acid open reading frames, simply by chance. This results in many false negatives during detection.
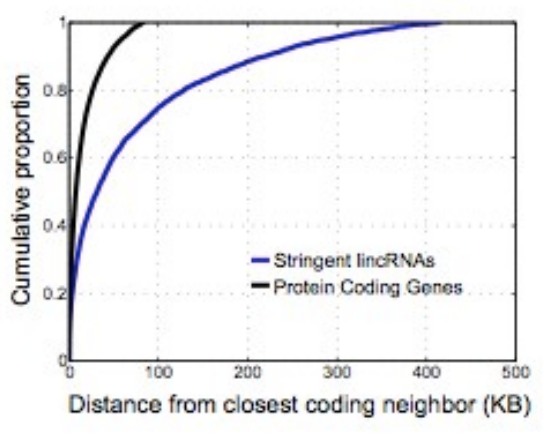
It has been found that many lincRNAs tend to neighbor developmental regions of the genome. They also tend to be lowly expressed compared to protein coding genes.
© source unknown. All rights reserved. This content is excluded from our Creative Commons license. For more information, see http://ocw.mit.edu/help/faq-fair-use/.
Example: p53
Independent validation: we use animal models, where one is a wild-type p53, andone is a knockout. We induce p53, then ask if lincRNAs turn on. 32 of 39 lincRNAs found associated with p53 were temporally induced upon turning on p53.
One RNA in particular sat next to a protein-coding gene in the p53 pathway. We tried to figure out if p53 bound to promoter and turned it on. To do this, we cloned the promoter of lincRNA, and asked does p53 turn it on? We IPed the p53 protein, to see if it associated with the lincRNA of the promoter. It turned out that lincRNA is directly related to p53 - p53 turns it on. P53 also turns genes off - certain lincRNAs act as a repressor.
From this example (and others), we start to see that RNAs usually have a protein partner
RNA can bring myriad of different proteins together, allowing the cell lots of diversity. In this way its similar to phosphorylation. RNAs bind to important chromatin complexes, and is required for reprogramming skin cells into stem cells.


